Introduction to BiocPkgToolsPlus
Kevin Rue-Albrecht
University of Oxfordkevin.rue-albrecht@imm.ox.ac.uk
22 October 2025
BiocPkgToolsPlus.RmdBasics
Install BiocPkgToolsPlus
R is an open-source statistical environment which can be
easily modified to enhance its functionality via packages. BiocPkgToolsPlus
is a R package available via the Bioconductor repository for packages.
R can be installed on any operating system from CRAN after which you can install
BiocPkgToolsPlus
by using the following commands in your R session:
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("BiocPkgToolsPlus")
## Check that you have a valid Bioconductor installation
BiocManager::valid()Required knowledge
BiocPkgToolsPlus is based on many other packages and in particular in those that have implemented the infrastructure needed for dealing with RNA-seq data (EDIT!). That is, packages like SummarizedExperiment (EDIT!).
If you are asking yourself the question “Where do I start using Bioconductor?” you might be interested in this blog post.
Asking for help
As package developers, we try to explain clearly how to use our
packages and in which order to use the functions. But R and
Bioconductor have a steep learning curve so it is critical
to learn where to ask for help. The blog post quoted above mentions some
but we would like to highlight the Bioconductor support site
as the main resource for getting help: remember to use the
BiocPkgToolsPlus tag and check the older
posts. Other alternatives are available such as creating GitHub
issues and tweeting. However, please note that if you want to receive
help you should adhere to the posting
guidelines. It is particularly critical that you provide a small
reproducible example and your session information so package developers
can track down the source of the error.
Citing BiocPkgToolsPlus
We hope that BiocPkgToolsPlus will be useful for your research. Please use the following information to cite the package and the overall approach. Thank you!
## Citation info
citation("BiocPkgToolsPlus")
#> To cite package 'BiocPkgToolsPlus' in publications use:
#>
#> kevinrue (2025). _Demonstration of a Bioconductor Package_.
#> doi:10.18129/B9.bioc.BiocPkgToolsPlus
#> <https://doi.org/10.18129/B9.bioc.BiocPkgToolsPlus>,
#> https://github.com/kevinrue/BiocPkgToolsPlus/BiocPkgToolsPlus - R
#> package version 0.99.0,
#> <http://www.bioconductor.org/packages/BiocPkgToolsPlus>.
#>
#> kevinrue (2025). "Demonstration of a Bioconductor Package."
#> _bioRxiv_. doi:10.1101/TODO <https://doi.org/10.1101/TODO>,
#> <https://www.biorxiv.org/content/10.1101/TODO>.
#>
#> To see these entries in BibTeX format, use 'print(<citation>,
#> bibtex=TRUE)', 'toBibtex(.)', or set
#> 'options(citation.bibtex.max=999)'.Quick start to using to BiocPkgToolsPlus
Use cases
Getting packages under a biocViews term
out <- get_packages_by_view("Spatial")
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
str(out)
#> chr [1:79] "alabaster.sfe" "Banksy" "BatchSVG" "betaHMM" "BulkSignalR" ...Getting packages at the intersection of multiple biocViews terms
out <- get_packages_by_views(c("Spatial", "SingleCell", "Sequencing"))
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
str(out)
#> chr [1:13] "DESpace" "knowYourCG" "miRspongeR" "mitology" "poem" ...Getting biocViews co-occurrence counts
library("dplyr")
out <- get_view_cooccurrences("Spatial")
out |> arrange(desc(value))
#> # A tibble: 65 × 2
#> package value
#> <chr> <int>
#> 1 Transcriptomics 51
#> 2 SingleCell 49
#> 3 GeneExpression 42
#> 4 DataImport 18
#> 5 RNASeq 15
#> 6 Clustering 14
#> 7 ImmunoOncology 10
#> 8 DataRepresentation 8
#> 9 CellBasedAssays 7
#> 10 QualityControl 7
#> # ℹ 55 more rowsThe output can be used to instantly produce a wordcloud with a choice
of packages, for instance
BiocStyle::CRANpkg("wordcloud2"):
library("wordcloud2")
wordcloud2(out, minRotation = -pi/2, maxRotation = pi/2, size = 0.5)Note that setting size to a value lower than 1 is a workaround sometimes needed to display the most frequent words, as per this thread.
Getting biocViews representation over time
The number of packages annotated by a single biocView term (or one of its children) can be obtained as follows:
out <- get_view_counts_over_time("Spatial")
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
#> Checking for Bioc Release Update
out
#> # A tibble: 40 × 2
#> date count
#> <date> <int>
#> 1 2006-01-01 0
#> 2 2006-07-01 0
#> 3 2007-01-01 0
#> 4 2007-07-01 0
#> 5 2008-01-01 0
#> 6 2008-07-01 0
#> 7 2009-01-01 0
#> 8 2009-07-01 0
#> 9 2010-01-01 0
#> 10 2010-07-01 0
#> # ℹ 30 more rowsThe output can be instantly plotted using
BiocStyle::CRANpkg("ggplot2"):
library("ggplot2")
ggplot(out, aes(date, count)) +
geom_point() +
labs(
title = "Spatial",
y = "Packages",
x = "Time"
)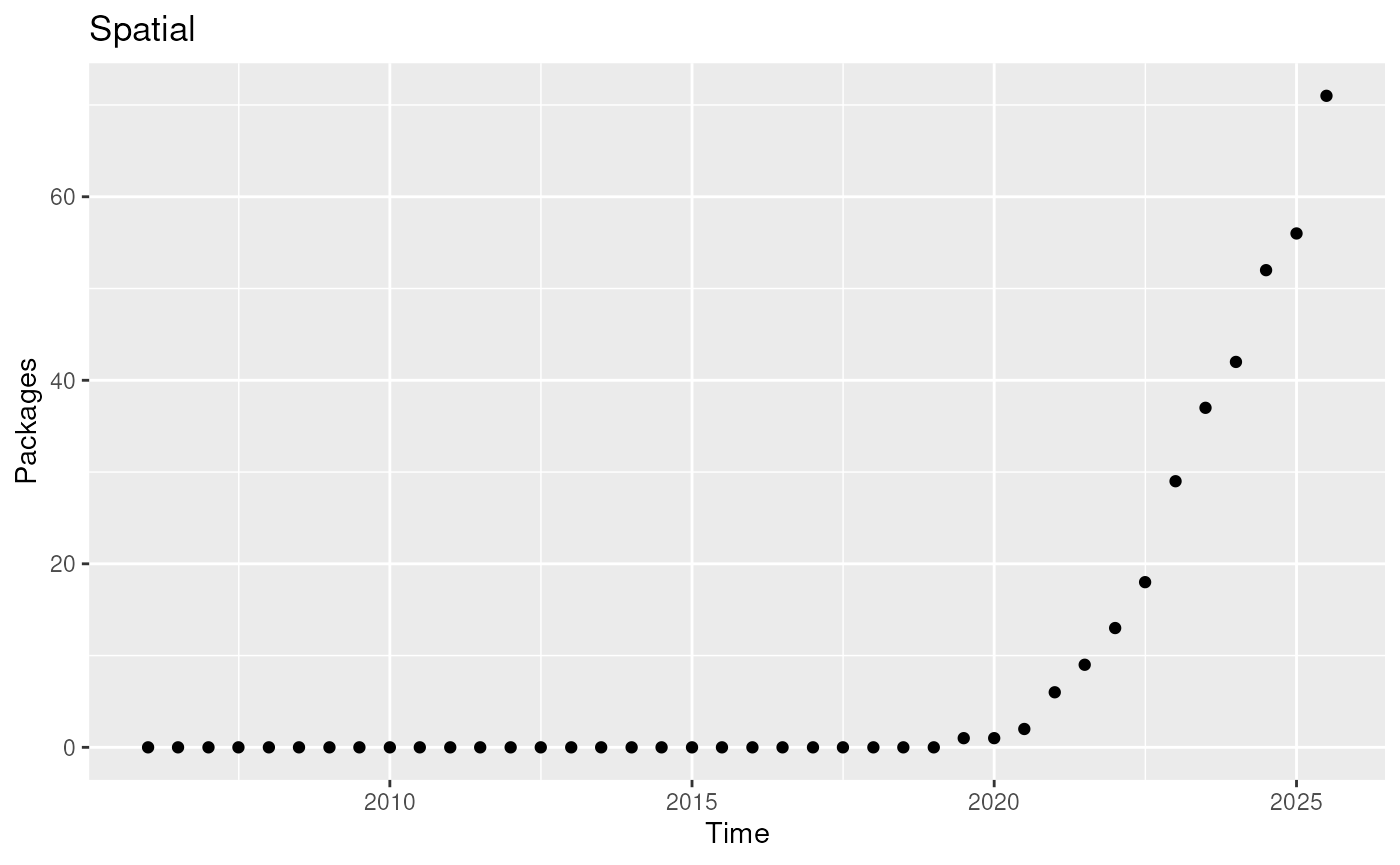
Count of ‘Spatial’ packages over time.
To get the counts for multiple biocViews terms, use
get_views_counts_over_time():
out <- get_views_counts_over_time(c("Spatial", "SingleCell"))
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
#> Checking for Bioc Release Update
out
#> # A tibble: 40 × 3
#> date Spatial SingleCell
#> <date> <int> <int>
#> 1 2006-01-01 0 0
#> 2 2006-07-01 0 0
#> 3 2007-01-01 0 0
#> 4 2007-07-01 0 0
#> 5 2008-01-01 0 0
#> 6 2008-07-01 0 0
#> 7 2009-01-01 0 1
#> 8 2009-07-01 0 1
#> 9 2010-01-01 0 1
#> 10 2010-07-01 0 1
#> # ℹ 30 more rowsWith a little bit more data wrangling, the output can also be plotted for all the terms at once, for instance:
library("tidyr")
library("ggplot2")
out|>
pivot_longer(!date) |>
ggplot(aes(date, value, colour = name)) +
geom_point() +
labs(
y = "Packages",
x = "Time",
colour = "biocView"
)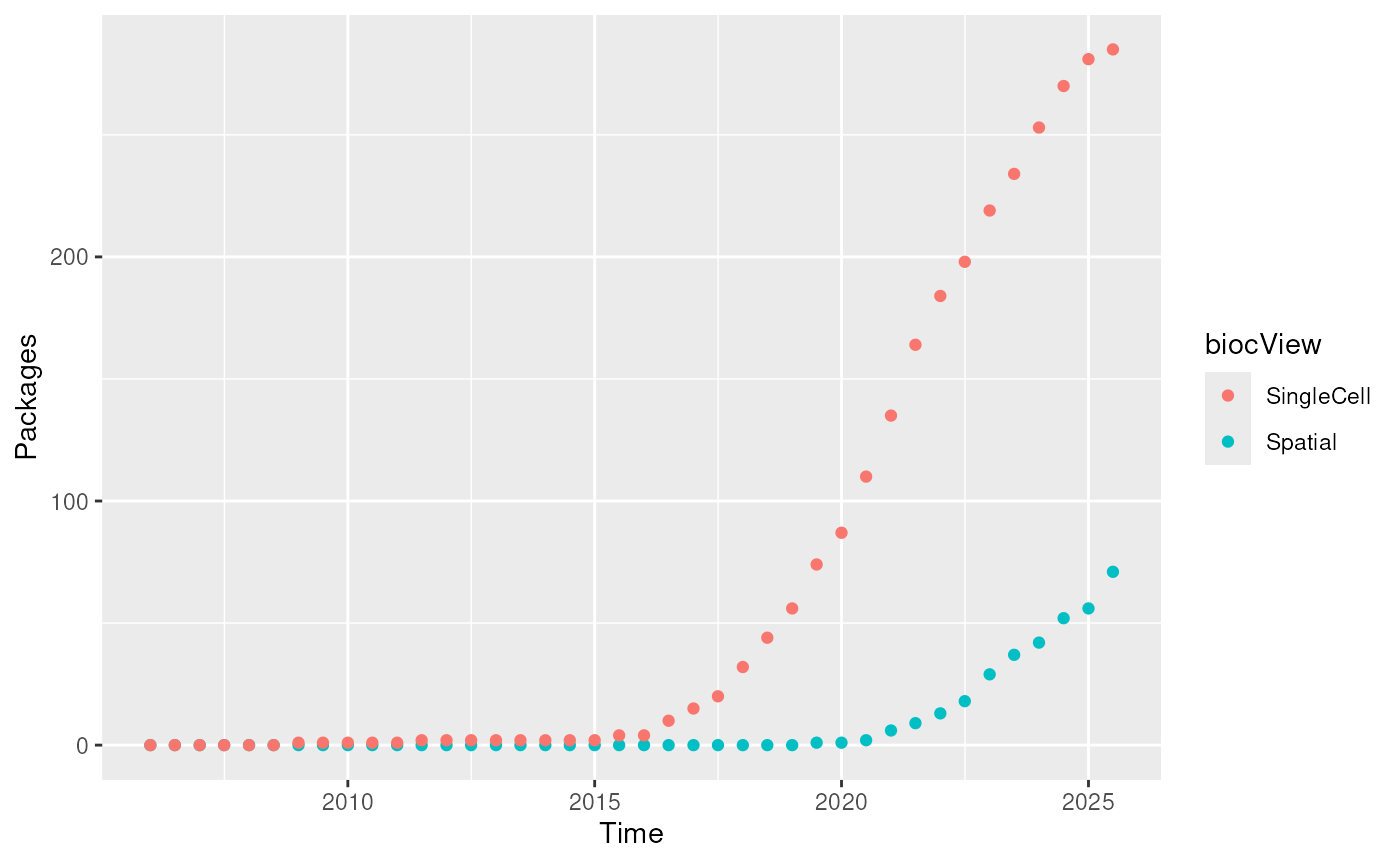
Count of ‘Spatial’ or ‘SingleCell’ packages over time.
Getting a package-by-biocView membership matrix
out <- get_view_membership_matrix()
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
out[1:5, 1:5]
#> BiocViews Software AnnotationData ExperimentData Workflow
#> a4 TRUE TRUE FALSE FALSE FALSE
#> a4Base TRUE TRUE FALSE FALSE FALSE
#> a4Classif TRUE TRUE FALSE FALSE FALSE
#> a4Core TRUE TRUE FALSE FALSE FALSE
#> a4Preproc TRUE TRUE FALSE FALSE FALSEGetting packages similar to a query package
out <- get_similar_packages("AUCell")
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
out
#> # A tibble: 2,352 × 2
#> package similarity
#> <chr> <dbl>
#> 1 UCell 0.975
#> 2 awst 0.969
#> 3 CellMixS 0.969
#> 4 CSOA 0.969
#> 5 EasyCellType 0.969
#> 6 HVP 0.969
#> 7 aggregateBioVar 0.963
#> 8 batchelor 0.963
#> 9 COTAN 0.963
#> 10 MAST 0.963
#> # ℹ 2,342 more rowsHere is an example of you can cite your package inside the vignette:
- BiocPkgToolsPlus (kevinrue, 2025)
Reproducibility
The BiocPkgToolsPlus package (kevinrue, 2025) was made possible thanks to:
- R (R Core Team, 2025)
- BiocStyle (Oleś, 2025)
- knitr (Xie, 2025)
- RefManageR (McLean, 2017)
- rmarkdown (Allaire, Xie, Dervieux, McPherson, Luraschi, Ushey, Atkins, Wickham, Cheng, Chang, and Iannone, 2025)
- sessioninfo (Wickham, Chang, Flight, Müller, and Hester, 2025)
- testthat (Wickham, 2011)
This package was developed using biocthis.
Code for creating the vignette
## Create the vignette
library("rmarkdown")
system.time(render("BiocPkgToolsPlus.Rmd", "BiocStyle::html_document"))
## Extract the R code
library("knitr")
knit("BiocPkgToolsPlus.Rmd", tangle = TRUE)Date the vignette was generated.
#> [1] "2025-10-22 08:40:39 UTC"Wallclock time spent generating the vignette.
#> Time difference of 24.331 secsR session information.
#> ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.5.1 (2025-06-13)
#> os Ubuntu 24.04.3 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz UTC
#> date 2025-10-22
#> pandoc 3.8.1 @ /usr/bin/ (via rmarkdown)
#> quarto 1.7.32 @ /usr/local/bin/quarto
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> backports 1.5.0 2024-05-23 [1] RSPM (R 4.5.0)
#> bibtex 0.5.1 2023-01-26 [1] RSPM (R 4.5.0)
#> Biobase 2.69.1 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocFileCache 2.99.6 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocGenerics 0.55.4 2025-10-17 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocManager 1.30.26 2025-06-05 [2] CRAN (R 4.5.1)
#> BiocPkgTools 1.27.12 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocPkgToolsPlus * 0.99.0 2025-10-22 [1] Bioconductor
#> BiocStyle * 2.37.1 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> biocViews * 1.77.4 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> bit 4.6.0 2025-03-06 [1] RSPM (R 4.5.0)
#> bit64 4.6.0-1 2025-01-16 [1] RSPM (R 4.5.0)
#> bitops 1.0-9 2024-10-03 [1] RSPM (R 4.5.0)
#> blob 1.2.4 2023-03-17 [1] RSPM (R 4.5.0)
#> bookdown 0.45 2025-10-03 [1] RSPM (R 4.5.0)
#> bslib 0.9.0 2025-01-30 [2] RSPM (R 4.5.0)
#> cachem 1.1.0 2024-05-16 [2] RSPM (R 4.5.0)
#> cli 3.6.5 2025-04-23 [2] RSPM (R 4.5.0)
#> curl 7.0.0 2025-08-19 [2] RSPM (R 4.5.0)
#> DBI 1.2.3 2024-06-02 [1] RSPM (R 4.5.0)
#> dbplyr 2.5.1 2025-09-10 [1] RSPM (R 4.5.0)
#> desc 1.4.3 2023-12-10 [2] RSPM (R 4.5.0)
#> digest 0.6.37 2024-08-19 [2] RSPM (R 4.5.0)
#> dplyr * 1.1.4 2023-11-17 [1] RSPM (R 4.5.0)
#> DT 0.34.0 2025-09-02 [1] RSPM (R 4.5.0)
#> evaluate 1.0.5 2025-08-27 [2] RSPM (R 4.5.0)
#> farver 2.1.2 2024-05-13 [1] RSPM (R 4.5.0)
#> fastmap 1.2.0 2024-05-15 [2] RSPM (R 4.5.0)
#> filelock 1.0.3 2023-12-11 [1] RSPM (R 4.5.0)
#> fs 1.6.6 2025-04-12 [2] RSPM (R 4.5.0)
#> generics 0.1.4 2025-05-09 [1] RSPM (R 4.5.0)
#> ggplot2 * 4.0.0 2025-09-11 [1] RSPM (R 4.5.0)
#> gh 1.5.0 2025-05-26 [2] RSPM (R 4.5.0)
#> glue 1.8.0 2024-09-30 [2] RSPM (R 4.5.0)
#> graph 1.87.0 2025-04-15 [1] Bioconductor 3.22 (R 4.5.0)
#> gtable 0.3.6 2024-10-25 [1] RSPM (R 4.5.0)
#> hms 1.1.4 2025-10-17 [1] RSPM (R 4.5.0)
#> htmltools 0.5.8.1 2024-04-04 [2] RSPM (R 4.5.0)
#> htmlwidgets 1.6.4 2023-12-06 [2] RSPM (R 4.5.0)
#> httr 1.4.7 2023-08-15 [1] RSPM (R 4.5.0)
#> httr2 1.2.1 2025-07-22 [2] RSPM (R 4.5.0)
#> igraph 2.2.0 2025-10-13 [1] RSPM (R 4.5.0)
#> jquerylib 0.1.4 2021-04-26 [2] RSPM (R 4.5.0)
#> jsonlite 2.0.0 2025-03-27 [2] RSPM (R 4.5.0)
#> knitr 1.50 2025-03-16 [2] RSPM (R 4.5.0)
#> labeling 0.4.3 2023-08-29 [1] RSPM (R 4.5.0)
#> lifecycle 1.0.4 2023-11-07 [2] RSPM (R 4.5.0)
#> lubridate 1.9.4 2024-12-08 [1] RSPM (R 4.5.0)
#> magrittr 2.0.4 2025-09-12 [2] RSPM (R 4.5.0)
#> memoise 2.0.1 2021-11-26 [2] RSPM (R 4.5.0)
#> pillar 1.11.1 2025-09-17 [2] RSPM (R 4.5.0)
#> pkgconfig 2.0.3 2019-09-22 [2] RSPM (R 4.5.0)
#> pkgdown 2.1.3 2025-05-25 [2] RSPM (R 4.5.0)
#> plyr 1.8.9 2023-10-02 [1] RSPM (R 4.5.0)
#> purrr 1.1.0 2025-07-10 [2] RSPM (R 4.5.0)
#> R6 2.6.1 2025-02-15 [2] RSPM (R 4.5.0)
#> ragg 1.5.0 2025-09-02 [2] RSPM (R 4.5.0)
#> rappdirs 0.3.3 2021-01-31 [2] RSPM (R 4.5.0)
#> RBGL 1.85.0 2025-04-15 [1] Bioconductor 3.22 (R 4.5.0)
#> RColorBrewer 1.1-3 2022-04-03 [1] RSPM (R 4.5.0)
#> Rcpp 1.1.0 2025-07-02 [2] RSPM (R 4.5.0)
#> RCurl 1.98-1.17 2025-03-22 [1] RSPM (R 4.5.0)
#> readr 2.1.5 2024-01-10 [1] RSPM (R 4.5.0)
#> RefManageR * 1.4.0 2022-09-30 [1] RSPM (R 4.5.0)
#> rlang 1.1.6 2025-04-11 [2] RSPM (R 4.5.0)
#> rmarkdown 2.30 2025-09-28 [2] RSPM (R 4.5.0)
#> RSQLite 2.4.3 2025-08-20 [1] RSPM (R 4.5.0)
#> RUnit 0.4.33.1 2025-06-17 [1] RSPM (R 4.5.0)
#> rvest 1.0.5 2025-08-29 [1] RSPM (R 4.5.0)
#> S7 0.2.0 2024-11-07 [1] RSPM (R 4.5.0)
#> sass 0.4.10 2025-04-11 [2] RSPM (R 4.5.0)
#> scales 1.4.0 2025-04-24 [1] RSPM (R 4.5.0)
#> sessioninfo * 1.2.3 2025-02-05 [2] RSPM (R 4.5.0)
#> stringi 1.8.7 2025-03-27 [2] RSPM (R 4.5.0)
#> stringr 1.5.2 2025-09-08 [2] RSPM (R 4.5.0)
#> systemfonts 1.3.1 2025-10-01 [2] RSPM (R 4.5.0)
#> textshaping 1.0.4 2025-10-10 [2] RSPM (R 4.5.0)
#> tibble 3.3.0 2025-06-08 [2] RSPM (R 4.5.0)
#> tidyr * 1.3.1 2024-01-24 [1] RSPM (R 4.5.0)
#> tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.5.0)
#> timechange 0.3.0 2024-01-18 [1] RSPM (R 4.5.0)
#> tzdb 0.5.0 2025-03-15 [1] RSPM (R 4.5.0)
#> utf8 1.2.6 2025-06-08 [2] RSPM (R 4.5.0)
#> vctrs 0.6.5 2023-12-01 [2] RSPM (R 4.5.0)
#> withr 3.0.2 2024-10-28 [2] RSPM (R 4.5.0)
#> wordcloud2 * 0.2.1 2018-01-03 [1] RSPM (R 4.5.0)
#> xfun 0.53 2025-08-19 [2] RSPM (R 4.5.0)
#> XML 3.99-0.19 2025-08-22 [1] RSPM (R 4.5.0)
#> xml2 1.4.0 2025-08-20 [2] RSPM (R 4.5.0)
#> yaml 2.3.10 2024-07-26 [2] RSPM (R 4.5.0)
#>
#> [1] /__w/_temp/Library
#> [2] /usr/local/lib/R/site-library
#> [3] /usr/local/lib/R/library
#> * ── Packages attached to the search path.
#>
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────Bibliography
This vignette was generated using BiocStyle (Oleś, 2025) with knitr (Xie, 2025) and rmarkdown (Allaire, Xie, Dervieux et al., 2025) running behind the scenes.
Citations made with RefManageR (McLean, 2017).
[1] J. Allaire, Y. Xie, C. Dervieux, et al. rmarkdown: Dynamic Documents for R. R package version 2.30. 2025. URL: https://github.com/rstudio/rmarkdown.
[2] kevinrue. Demonstration of a Bioconductor Package. https://github.com/kevinrue/BiocPkgToolsPlus/BiocPkgToolsPlus - R package version 0.99.0. 2025. DOI: 10.18129/B9.bioc.BiocPkgToolsPlus. URL: http://www.bioconductor.org/packages/BiocPkgToolsPlus.
[3] M. W. McLean. “RefManageR: Import and Manage BibTeX and BibLaTeX References in R”. In: The Journal of Open Source Software (2017). DOI: 10.21105/joss.00338.
[4] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.37.1. 2025. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
[5] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2025. URL: https://www.R-project.org/.
[6] H. Wickham. “testthat: Get Started with Testing”. In: The R Journal 3 (2011), pp. 5–10. URL: https://journal.r-project.org/archive/2011-1/RJournal_2011-1_Wickham.pdf.
[7] H. Wickham, W. Chang, R. Flight, et al. sessioninfo: R Session Information. R package version 1.2.3. 2025. URL: https://github.com/r-lib/sessioninfo#readme.
[8] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.50. 2025. URL: https://yihui.org/knitr/.