Optimisations
Kevin Rue-Albrecht
University of Oxfordkevin.rue-albrecht@imm.ox.ac.uk
22 October 2025
Optimisations.RmdLibraries
Attach packages used in this vignettes.
library("BiocPkgTools")
#> Loading required package: htmlwidgets
library("BiocPkgToolsPlus")
#> Loading required package: biocViewsAvoiding repeated calls to biocPkgList()
To save some typing, get_packages_by_view() calls
biocPkgList() internally if pkg_list is
NULL (the default). This is convenient for one-off calls,
but slow and verbose for multiple calls.
If you are calling get_packages_by_view() multiple
times, it is more efficient to call biocPkgList() once and
pass the result to get_packages_by_view().
pkg_list <- biocPkgList()
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://p3m.dev/cran/__linux__/noble/latest
out1 <- get_packages_by_view("Spatial", pkg_list = pkg_list)
out2 <- get_packages_by_view("SingleCell", pkg_list = pkg_list)Wordclouds
The ‘Getting started’ vignette demonstrated a single method for producing a wordcloud. This section offers alternatives.
Let’s start by getting the count of co-occurrences for each biocViews term with a query term, obtained as follows:
library("dplyr")
out <- get_view_cooccurrences(view = "Spatial", pkg_list = pkg_list)
out |> arrange(desc(value))
#> # A tibble: 65 × 2
#> package value
#> <chr> <int>
#> 1 Transcriptomics 51
#> 2 SingleCell 49
#> 3 GeneExpression 42
#> 4 DataImport 18
#> 5 RNASeq 15
#> 6 Clustering 14
#> 7 ImmunoOncology 10
#> 8 DataRepresentation 8
#> 9 CellBasedAssays 7
#> 10 QualityControl 7
#> # ℹ 55 more rowsIn contrast to the interactive workclouds produced by
BiocStyle::CRANpkg("wordcloud2"), the
BiocStyle::CRANpkg("wordcloud") package can be used to
produce static wordclouds, with a little bit more work:
library(wordcloud)
library(RColorBrewer)
set.seed(1)
wordcloud(
words = out$package,
freq = out$value,
colors = brewer.pal(n = 8, name = "Dark2"),
random.color = TRUE,
rot.per=.2,
scale=c(3, .5)
)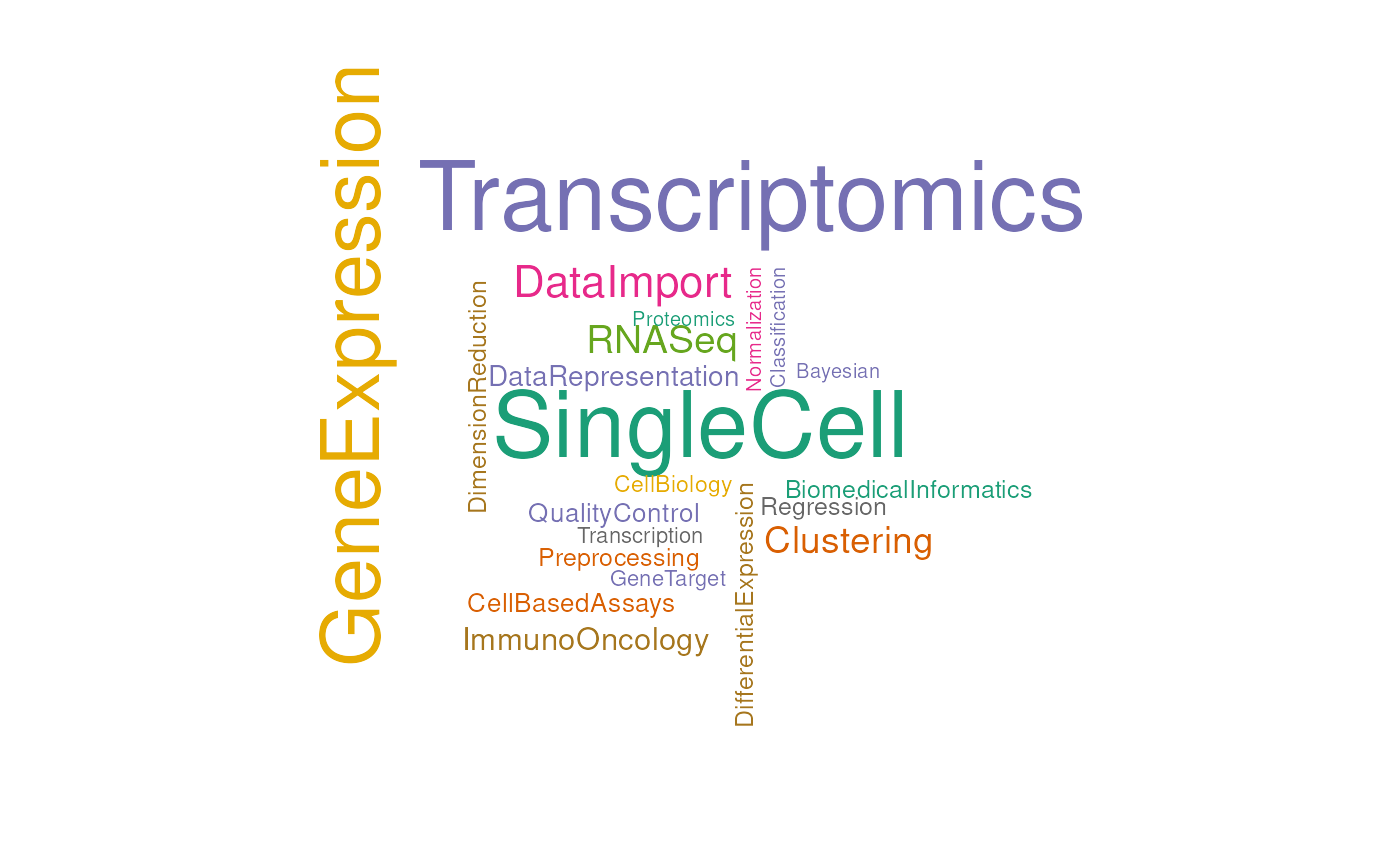
Wordcloud.
Reproducibility
The BiocPkgToolsPlus package (kevinrue, 2025) was made possible thanks to:
- R (R Core Team, 2025)
- BiocStyle (Oleś, 2025)
- knitr (Xie, 2025)
- RefManageR (McLean, 2017)
- rmarkdown (Allaire, Xie, Dervieux, McPherson, Luraschi, Ushey, Atkins, Wickham, Cheng, Chang, and Iannone, 2025)
- sessioninfo (Wickham, Chang, Flight, Müller, and Hester, 2025)
- testthat (Wickham, 2011)
This package was developed using biocthis.
Code for creating the vignette
## Create the vignette
library("rmarkdown")
system.time(render("BiocPkgToolsPlus.Rmd", "BiocStyle::html_document"))
## Extract the R code
library("knitr")
knit("BiocPkgToolsPlus.Rmd", tangle = TRUE)Date the vignette was generated.
#> [1] "2025-10-22 08:40:46 UTC"Wallclock time spent generating the vignette.
#> Time difference of 3.99 secsR session information.
#> ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.5.1 (2025-06-13)
#> os Ubuntu 24.04.3 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz UTC
#> date 2025-10-22
#> pandoc 3.8.1 @ /usr/bin/ (via rmarkdown)
#> quarto 1.7.32 @ /usr/local/bin/quarto
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> backports 1.5.0 2024-05-23 [1] RSPM (R 4.5.0)
#> bibtex 0.5.1 2023-01-26 [1] RSPM (R 4.5.0)
#> Biobase 2.69.1 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocFileCache 2.99.6 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocGenerics 0.55.4 2025-10-17 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocManager 1.30.26 2025-06-05 [2] CRAN (R 4.5.1)
#> BiocPkgTools * 1.27.12 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> BiocPkgToolsPlus * 0.99.0 2025-10-22 [1] Bioconductor
#> BiocStyle * 2.37.1 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> biocViews * 1.77.4 2025-10-07 [1] Bioconductor 3.22 (R 4.5.1)
#> bit 4.6.0 2025-03-06 [1] RSPM (R 4.5.0)
#> bit64 4.6.0-1 2025-01-16 [1] RSPM (R 4.5.0)
#> bitops 1.0-9 2024-10-03 [1] RSPM (R 4.5.0)
#> blob 1.2.4 2023-03-17 [1] RSPM (R 4.5.0)
#> bookdown 0.45 2025-10-03 [1] RSPM (R 4.5.0)
#> bslib 0.9.0 2025-01-30 [2] RSPM (R 4.5.0)
#> cachem 1.1.0 2024-05-16 [2] RSPM (R 4.5.0)
#> cli 3.6.5 2025-04-23 [2] RSPM (R 4.5.0)
#> curl 7.0.0 2025-08-19 [2] RSPM (R 4.5.0)
#> DBI 1.2.3 2024-06-02 [1] RSPM (R 4.5.0)
#> dbplyr 2.5.1 2025-09-10 [1] RSPM (R 4.5.0)
#> desc 1.4.3 2023-12-10 [2] RSPM (R 4.5.0)
#> digest 0.6.37 2024-08-19 [2] RSPM (R 4.5.0)
#> dplyr * 1.1.4 2023-11-17 [1] RSPM (R 4.5.0)
#> DT 0.34.0 2025-09-02 [1] RSPM (R 4.5.0)
#> evaluate 1.0.5 2025-08-27 [2] RSPM (R 4.5.0)
#> fastmap 1.2.0 2024-05-15 [2] RSPM (R 4.5.0)
#> filelock 1.0.3 2023-12-11 [1] RSPM (R 4.5.0)
#> fs 1.6.6 2025-04-12 [2] RSPM (R 4.5.0)
#> generics 0.1.4 2025-05-09 [1] RSPM (R 4.5.0)
#> gh 1.5.0 2025-05-26 [2] RSPM (R 4.5.0)
#> glue 1.8.0 2024-09-30 [2] RSPM (R 4.5.0)
#> graph 1.87.0 2025-04-15 [1] Bioconductor 3.22 (R 4.5.0)
#> hms 1.1.4 2025-10-17 [1] RSPM (R 4.5.0)
#> htmltools 0.5.8.1 2024-04-04 [2] RSPM (R 4.5.0)
#> htmlwidgets * 1.6.4 2023-12-06 [2] RSPM (R 4.5.0)
#> httr 1.4.7 2023-08-15 [1] RSPM (R 4.5.0)
#> httr2 1.2.1 2025-07-22 [2] RSPM (R 4.5.0)
#> igraph 2.2.0 2025-10-13 [1] RSPM (R 4.5.0)
#> jquerylib 0.1.4 2021-04-26 [2] RSPM (R 4.5.0)
#> jsonlite 2.0.0 2025-03-27 [2] RSPM (R 4.5.0)
#> knitr 1.50 2025-03-16 [2] RSPM (R 4.5.0)
#> lifecycle 1.0.4 2023-11-07 [2] RSPM (R 4.5.0)
#> lubridate 1.9.4 2024-12-08 [1] RSPM (R 4.5.0)
#> magrittr 2.0.4 2025-09-12 [2] RSPM (R 4.5.0)
#> memoise 2.0.1 2021-11-26 [2] RSPM (R 4.5.0)
#> pillar 1.11.1 2025-09-17 [2] RSPM (R 4.5.0)
#> pkgconfig 2.0.3 2019-09-22 [2] RSPM (R 4.5.0)
#> pkgdown 2.1.3 2025-05-25 [2] RSPM (R 4.5.0)
#> plyr 1.8.9 2023-10-02 [1] RSPM (R 4.5.0)
#> purrr 1.1.0 2025-07-10 [2] RSPM (R 4.5.0)
#> R6 2.6.1 2025-02-15 [2] RSPM (R 4.5.0)
#> ragg 1.5.0 2025-09-02 [2] RSPM (R 4.5.0)
#> rappdirs 0.3.3 2021-01-31 [2] RSPM (R 4.5.0)
#> RBGL 1.85.0 2025-04-15 [1] Bioconductor 3.22 (R 4.5.0)
#> RColorBrewer * 1.1-3 2022-04-03 [1] RSPM (R 4.5.0)
#> Rcpp 1.1.0 2025-07-02 [2] RSPM (R 4.5.0)
#> RCurl 1.98-1.17 2025-03-22 [1] RSPM (R 4.5.0)
#> readr 2.1.5 2024-01-10 [1] RSPM (R 4.5.0)
#> RefManageR * 1.4.0 2022-09-30 [1] RSPM (R 4.5.0)
#> rlang 1.1.6 2025-04-11 [2] RSPM (R 4.5.0)
#> rmarkdown 2.30 2025-09-28 [2] RSPM (R 4.5.0)
#> RSQLite 2.4.3 2025-08-20 [1] RSPM (R 4.5.0)
#> RUnit 0.4.33.1 2025-06-17 [1] RSPM (R 4.5.0)
#> rvest 1.0.5 2025-08-29 [1] RSPM (R 4.5.0)
#> sass 0.4.10 2025-04-11 [2] RSPM (R 4.5.0)
#> sessioninfo * 1.2.3 2025-02-05 [2] RSPM (R 4.5.0)
#> stringi 1.8.7 2025-03-27 [2] RSPM (R 4.5.0)
#> stringr 1.5.2 2025-09-08 [2] RSPM (R 4.5.0)
#> systemfonts 1.3.1 2025-10-01 [2] RSPM (R 4.5.0)
#> textshaping 1.0.4 2025-10-10 [2] RSPM (R 4.5.0)
#> tibble 3.3.0 2025-06-08 [2] RSPM (R 4.5.0)
#> tidyr 1.3.1 2024-01-24 [1] RSPM (R 4.5.0)
#> tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.5.0)
#> timechange 0.3.0 2024-01-18 [1] RSPM (R 4.5.0)
#> tzdb 0.5.0 2025-03-15 [1] RSPM (R 4.5.0)
#> utf8 1.2.6 2025-06-08 [2] RSPM (R 4.5.0)
#> vctrs 0.6.5 2023-12-01 [2] RSPM (R 4.5.0)
#> wordcloud * 2.6 2018-08-24 [1] RSPM (R 4.5.0)
#> xfun 0.53 2025-08-19 [2] RSPM (R 4.5.0)
#> XML 3.99-0.19 2025-08-22 [1] RSPM (R 4.5.0)
#> xml2 1.4.0 2025-08-20 [2] RSPM (R 4.5.0)
#> yaml 2.3.10 2024-07-26 [2] RSPM (R 4.5.0)
#>
#> [1] /__w/_temp/Library
#> [2] /usr/local/lib/R/site-library
#> [3] /usr/local/lib/R/library
#> * ── Packages attached to the search path.
#>
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────Bibliography
This vignette was generated using BiocStyle (Oleś, 2025) with knitr (Xie, 2025) and rmarkdown (Allaire, Xie, Dervieux et al., 2025) running behind the scenes.
Citations made with RefManageR (McLean, 2017).
[1] J. Allaire, Y. Xie, C. Dervieux, et al. rmarkdown: Dynamic Documents for R. R package version 2.30. 2025. URL: https://github.com/rstudio/rmarkdown.
[2] kevinrue. Demonstration of a Bioconductor Package. https://github.com/kevinrue/BiocPkgToolsPlus/BiocPkgToolsPlus - R package version 0.99.0. 2025. DOI: 10.18129/B9.bioc.BiocPkgToolsPlus. URL: http://www.bioconductor.org/packages/BiocPkgToolsPlus.
[3] M. W. McLean. “RefManageR: Import and Manage BibTeX and BibLaTeX References in R”. In: The Journal of Open Source Software (2017). DOI: 10.21105/joss.00338.
[4] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.37.1. 2025. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
[5] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2025. URL: https://www.R-project.org/.
[6] H. Wickham. “testthat: Get Started with Testing”. In: The R Journal 3 (2011), pp. 5–10. URL: https://journal.r-project.org/archive/2011-1/RJournal_2011-1_Wickham.pdf.
[7] H. Wickham, W. Chang, R. Flight, et al. sessioninfo: R Session Information. R package version 1.2.3. 2025. URL: https://github.com/r-lib/sessioninfo#readme.
[8] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.50. 2025. URL: https://yihui.org/knitr/.