BioC2020
Bioconductor 2020 conference
2021-02-21 23:30:53
Parameters
| Parameter | Value |
|---|---|
| hashtag | #BioC2020 |
| start_day | 2020-07-27 |
| end_day | 2020-07-31 |
| timezone | America/New_York |
| theme | theme_light |
| accent | #0092ac |
| accent2 | #87b13f |
| kcore | 2 |
| topics_k | 6 |
| bigram_filter | 3 |
| fixed | TRUE |
| seed | 1 |
1 Introduction
An analysis of tweets from the #BioC2020 hashtag for the Bioconductor 2020 conference, 27-31 July 2020 online (originally planned in Boston, USA).
A total of 1946 tweets from 381 users were collected using the rtweet R package.
The latest tweet was recorded at 2020-08-01 07:53:54 (Etc/UTC).
2 Timeline
2.1 Tweets by day
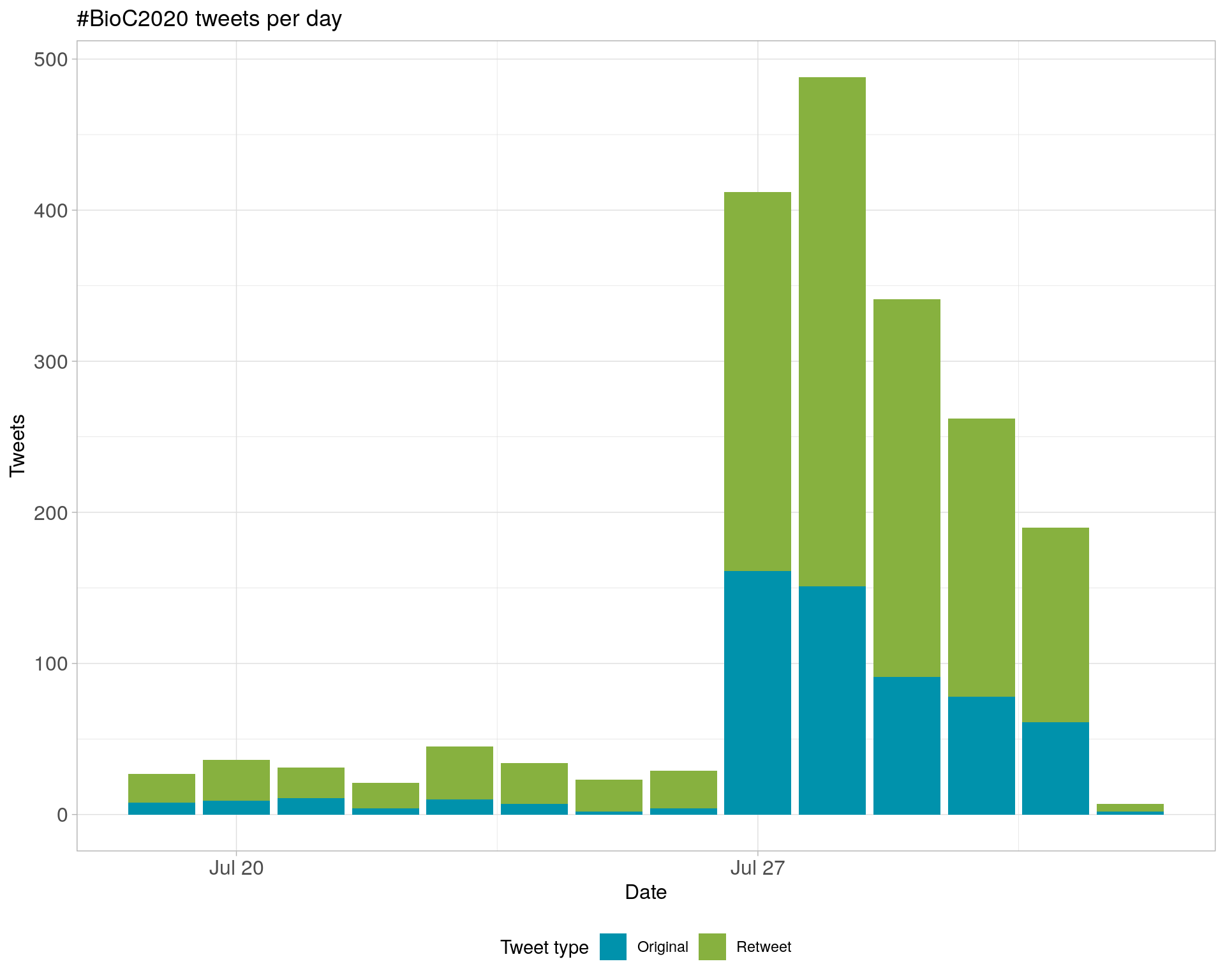
2.2 Tweets by day and time
Filtered for dates 2020-07-27 - 2020-07-31 in the America/New_York timezone.

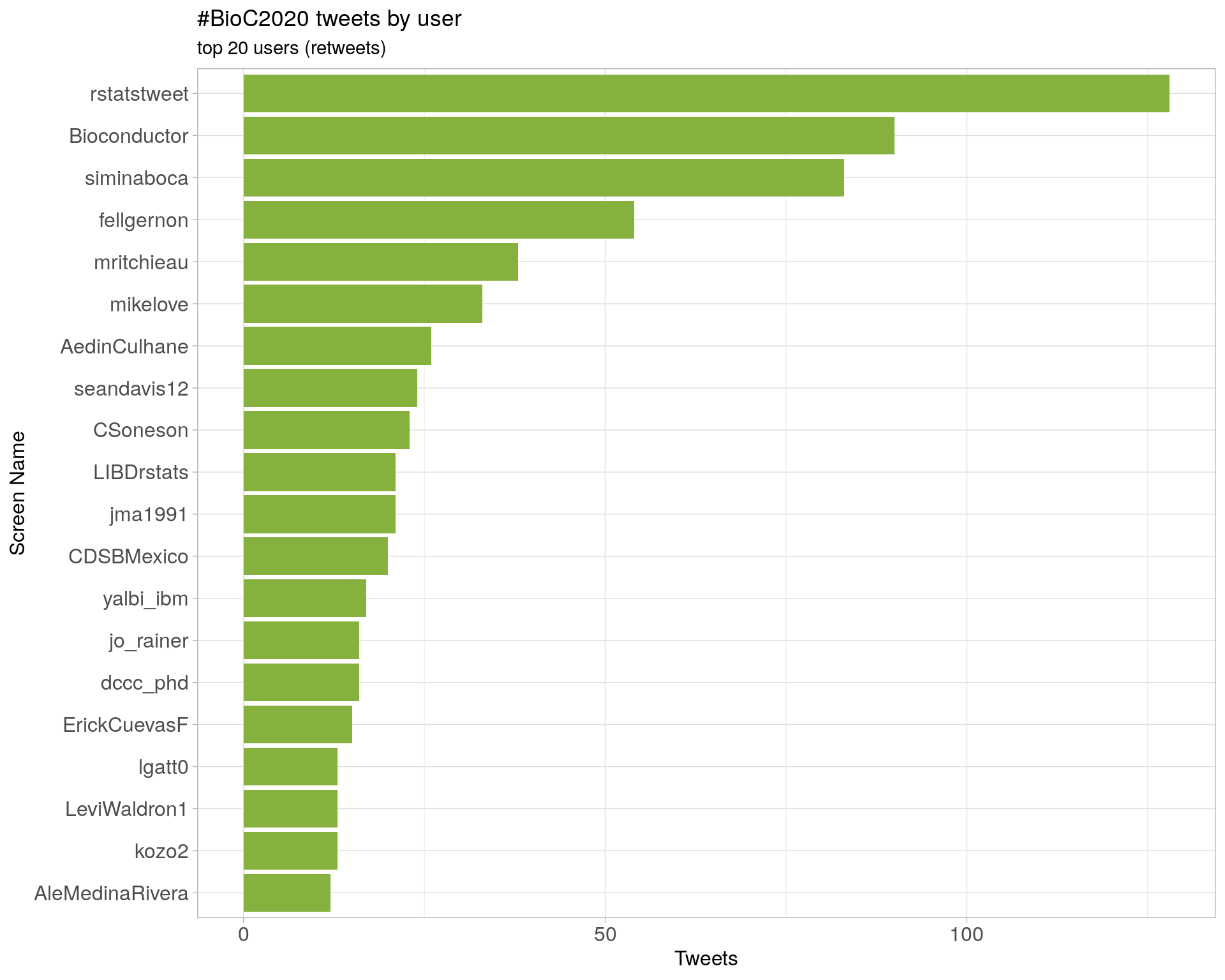
3 Users
3.1 Top tweeters
Overall

Original

Retweets
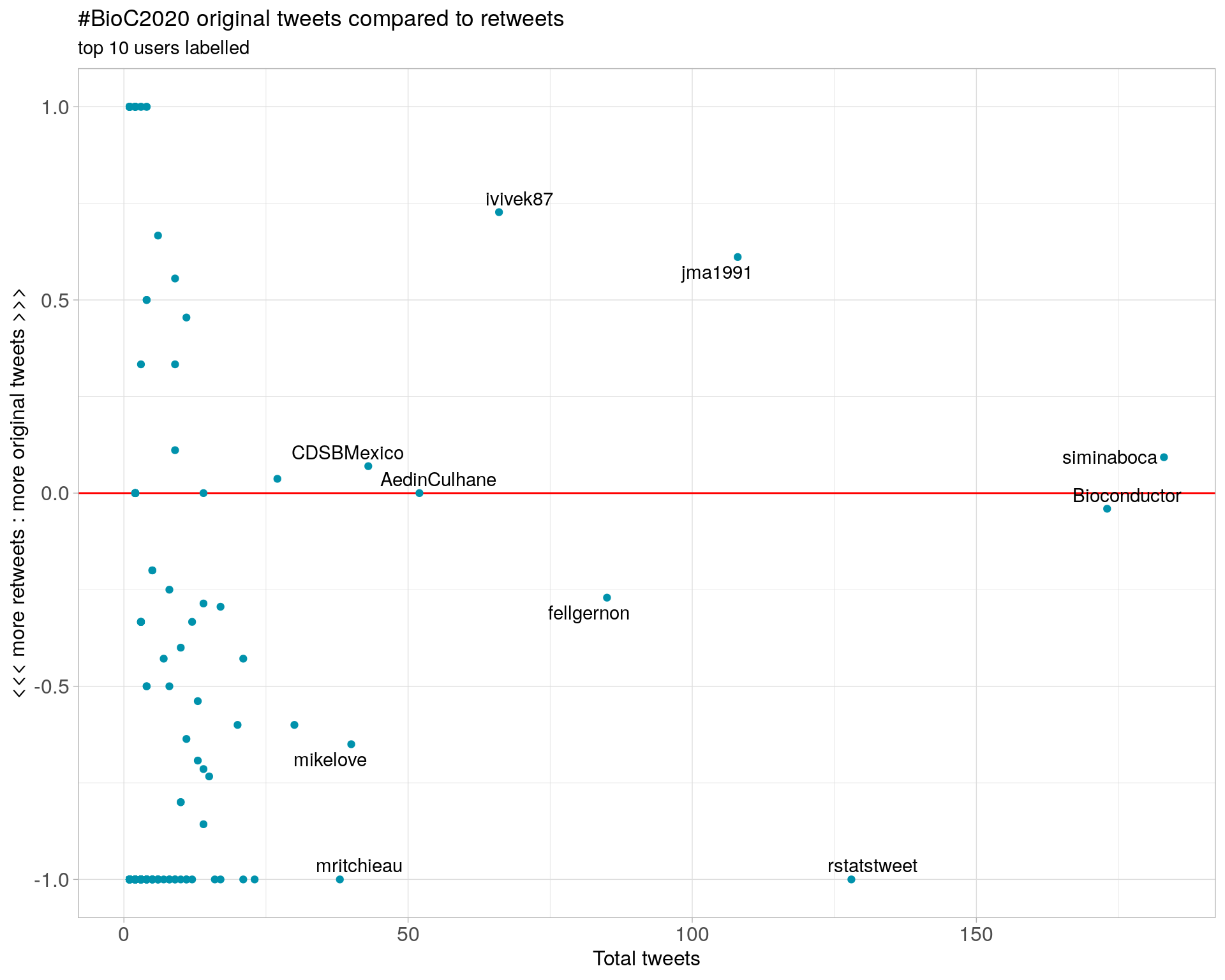
3.2 Retweet proportion
3.3 Top tweeters timeline

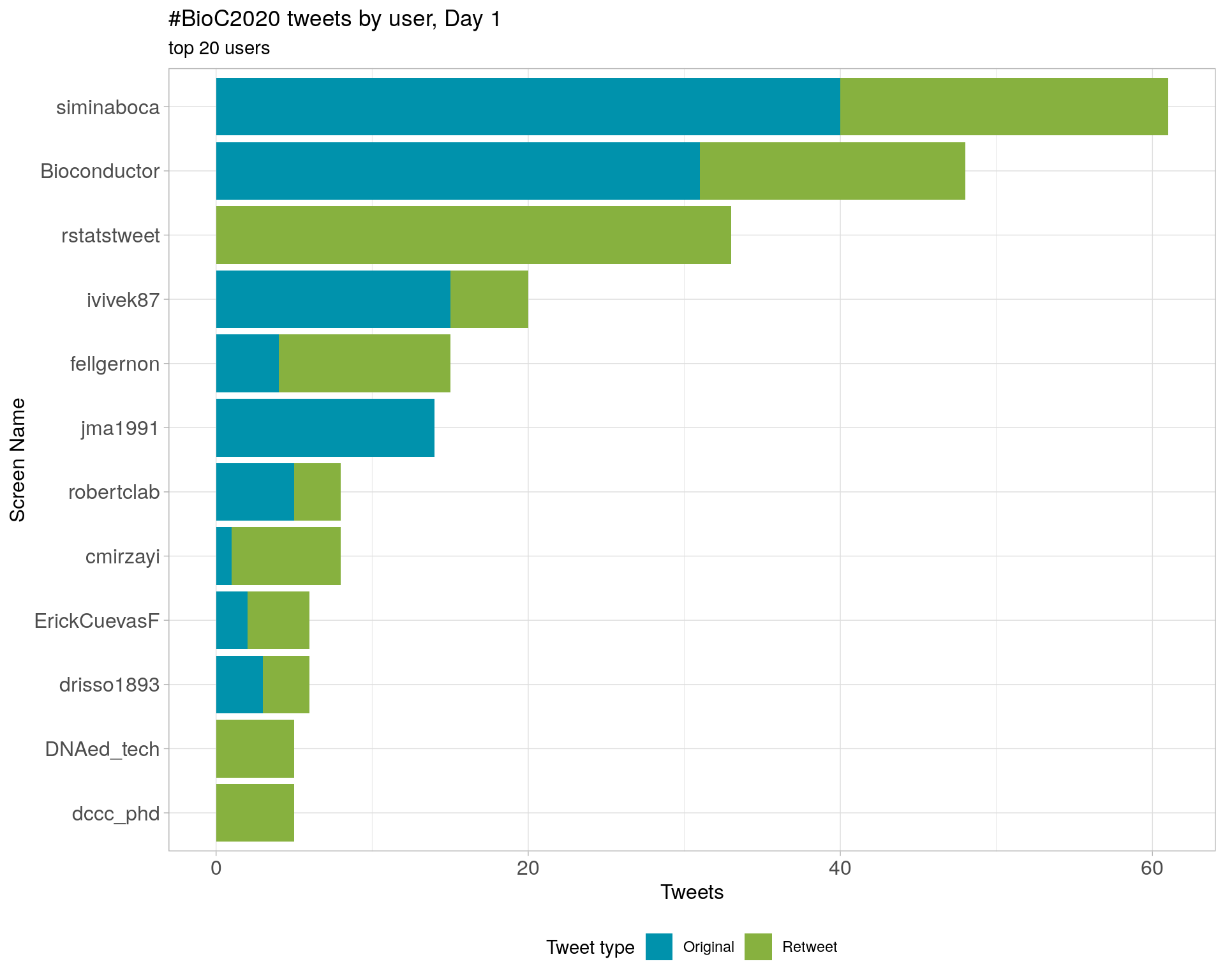
3.4 Top tweeters by day
Overall
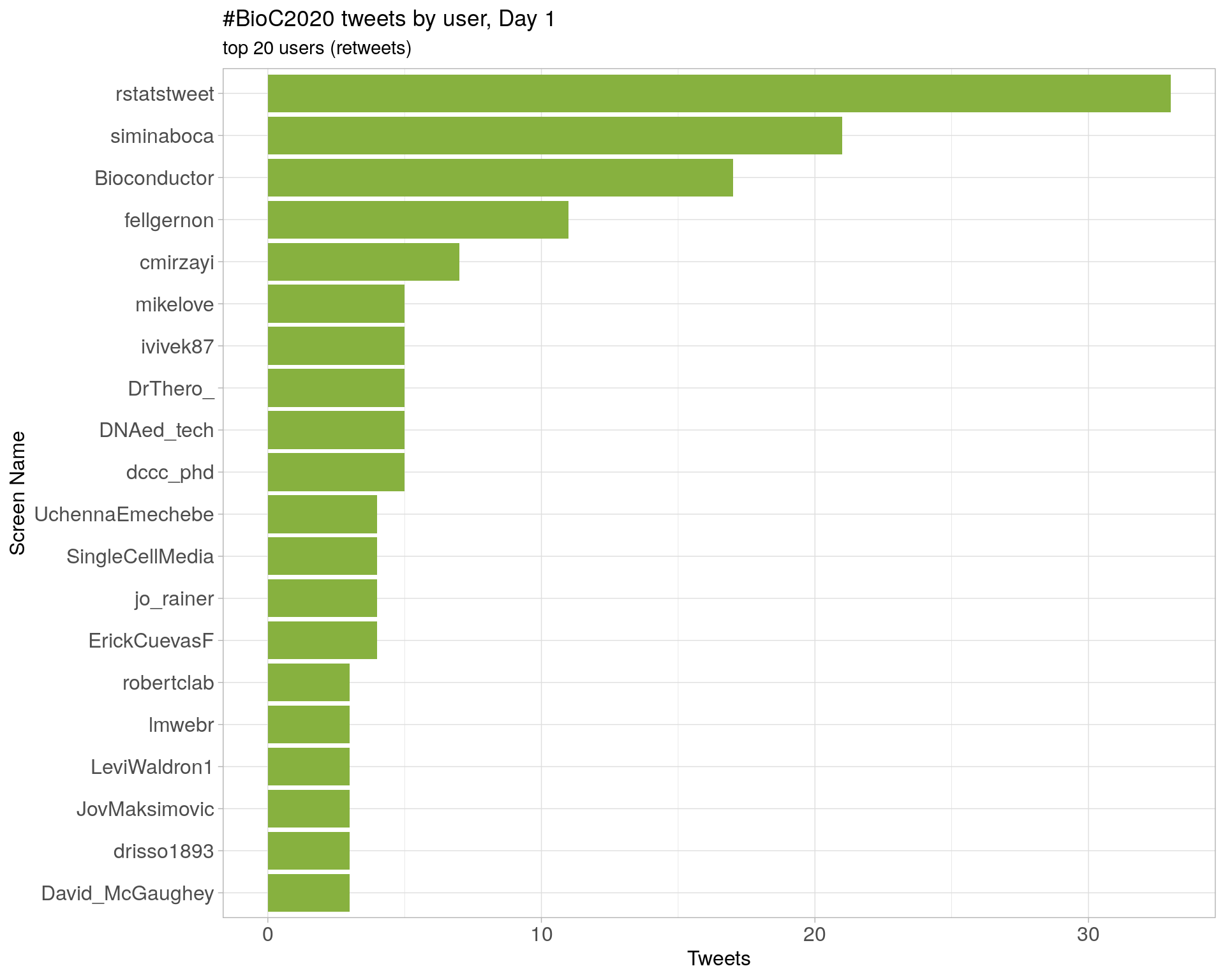
Day 1
Day 2
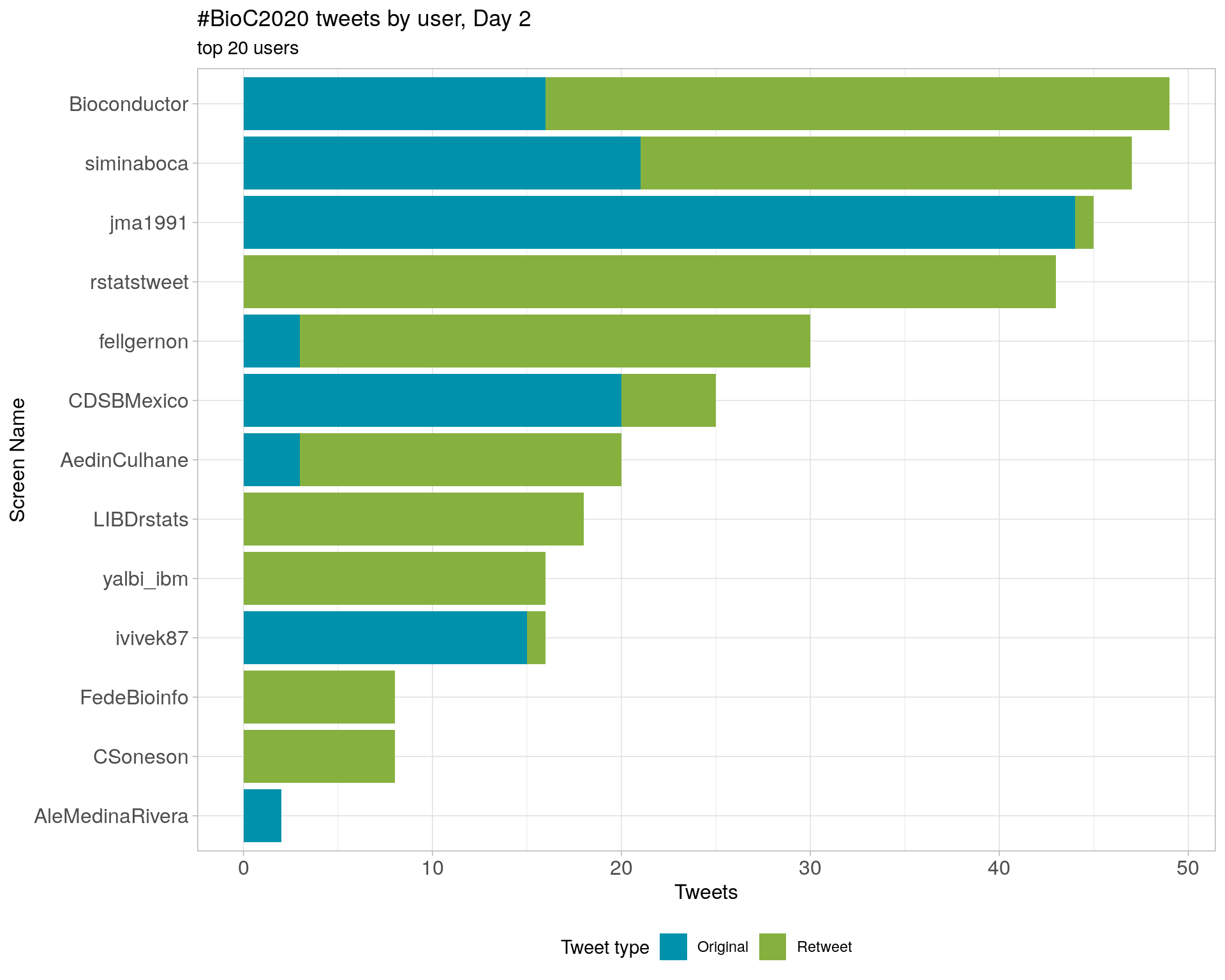
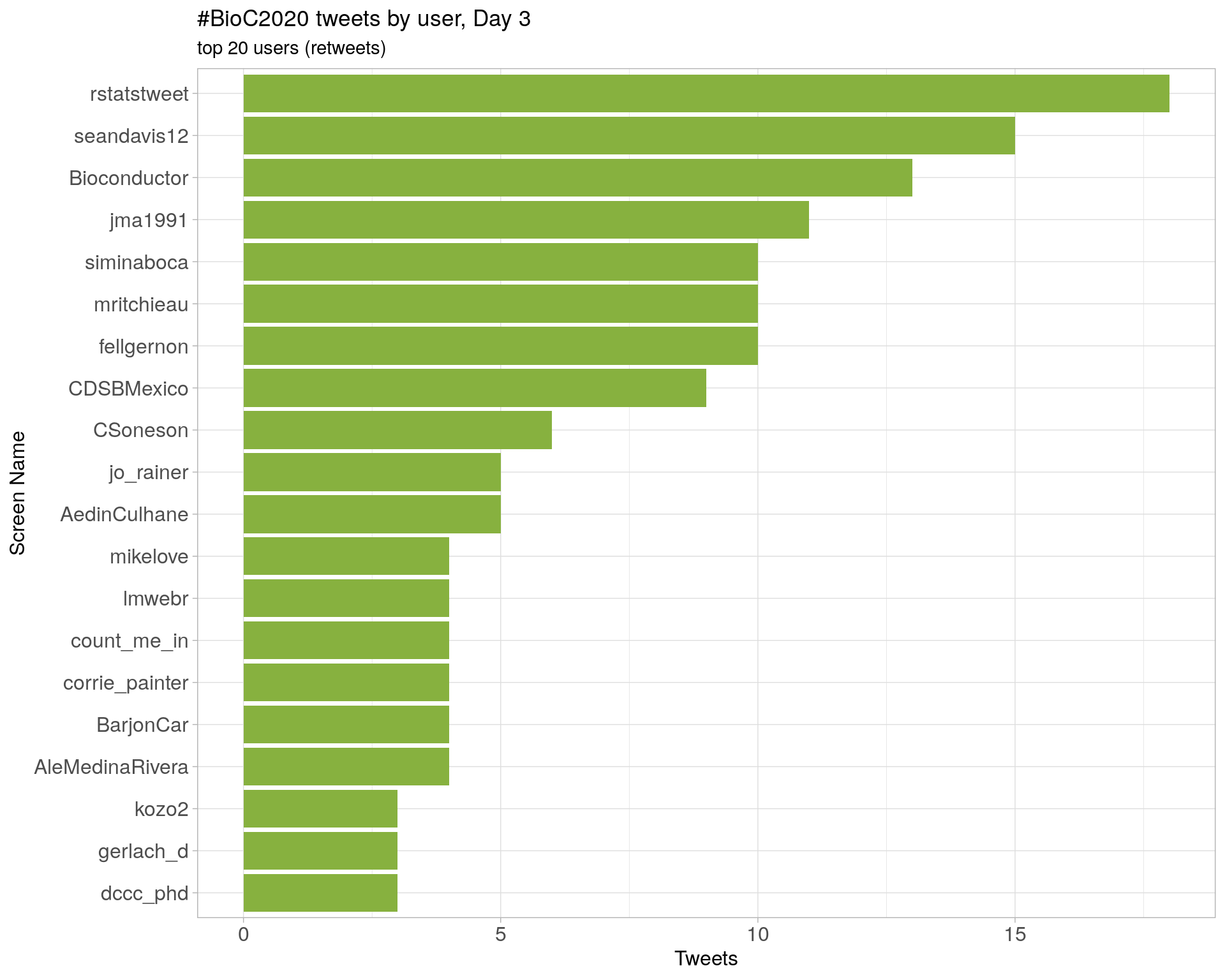
Day 3

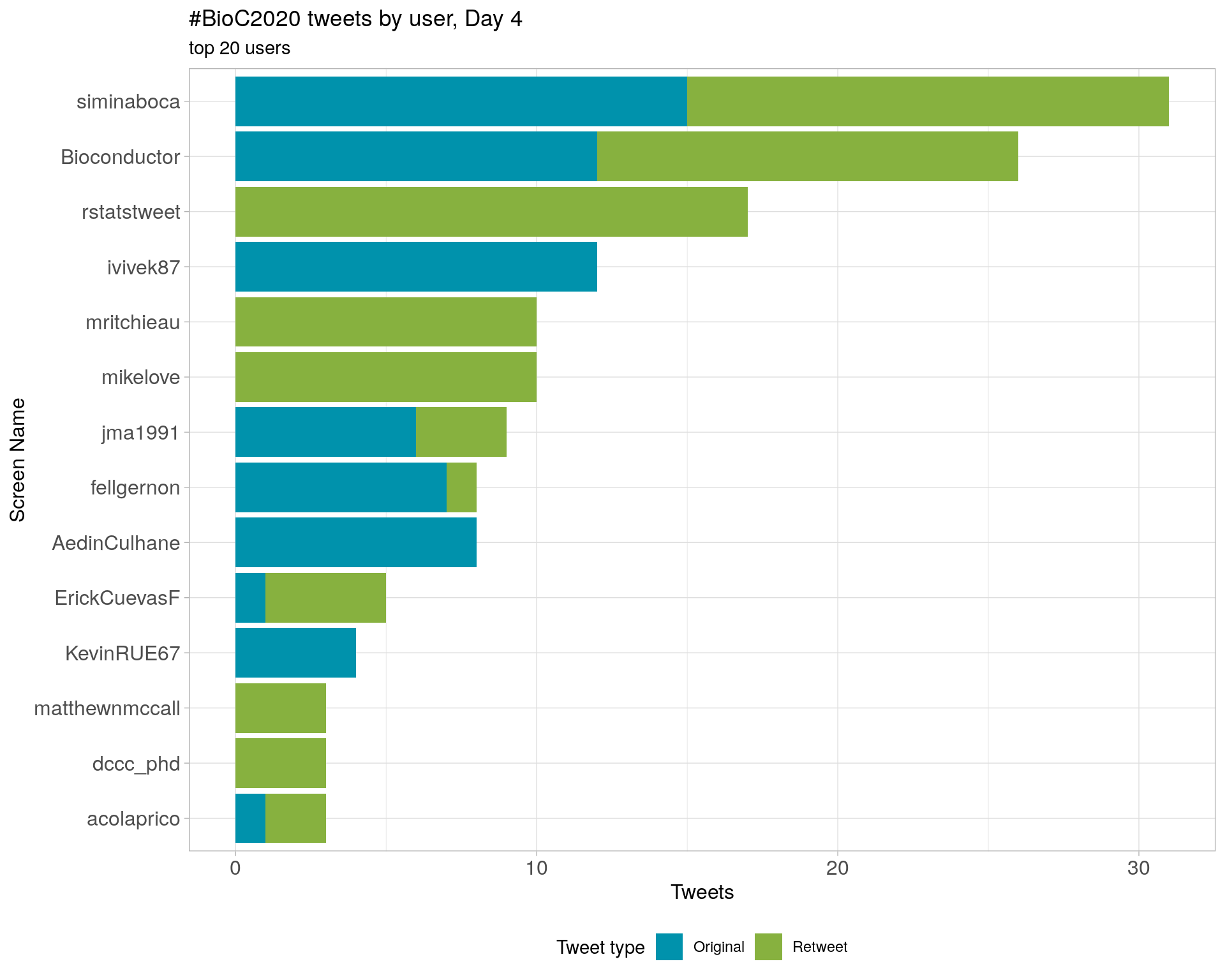
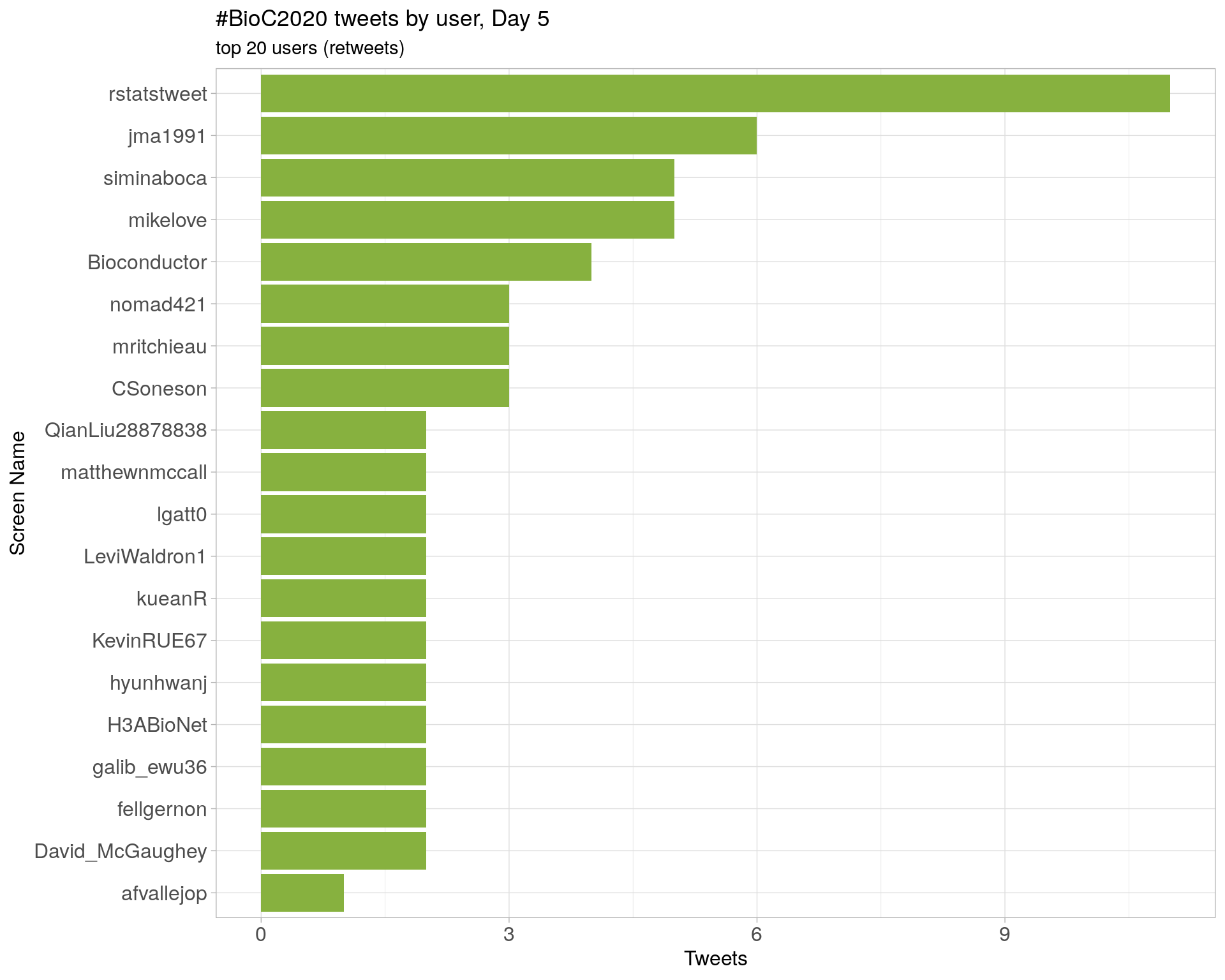
Day 4
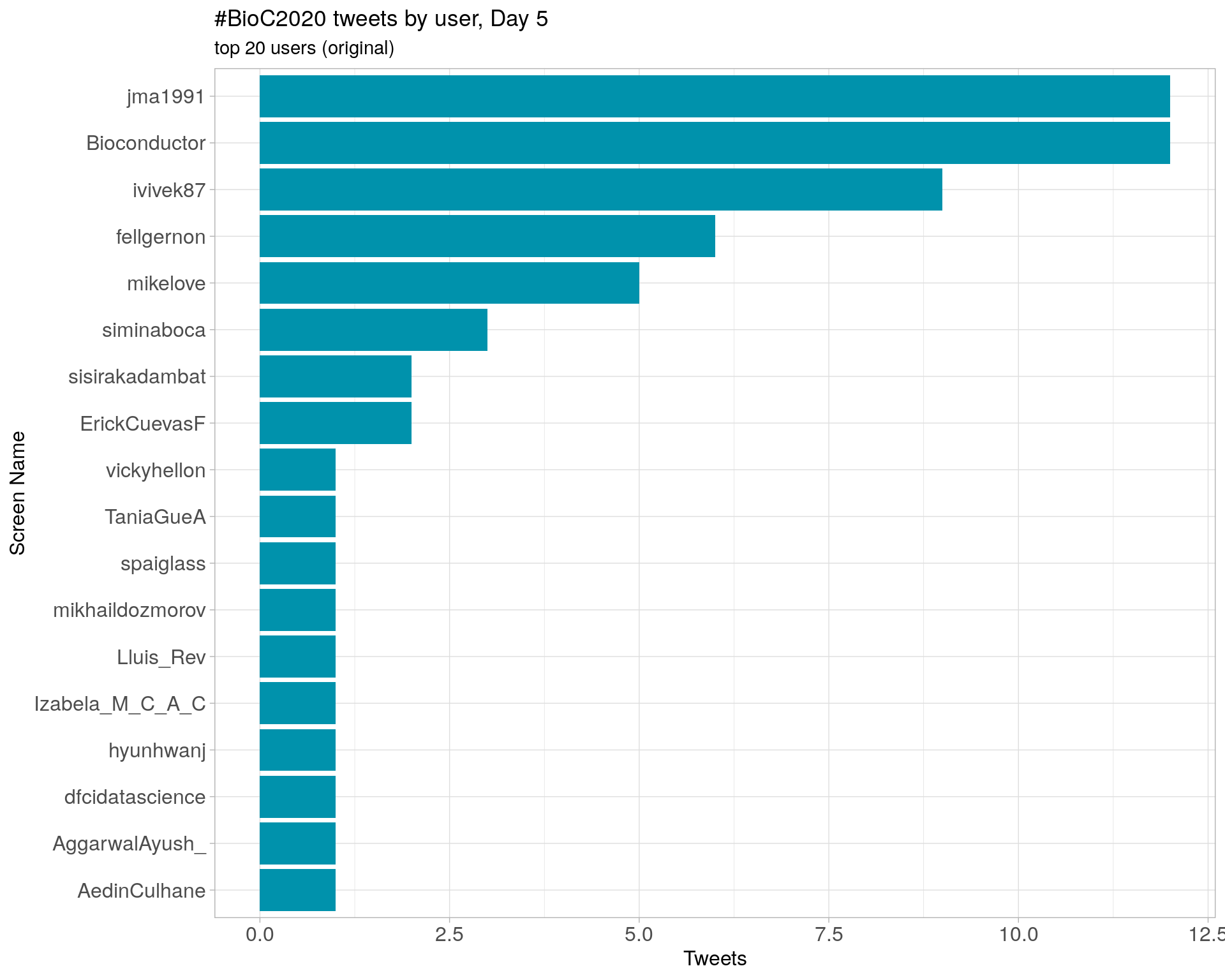
Day 5

Original
Day 1

Day 2
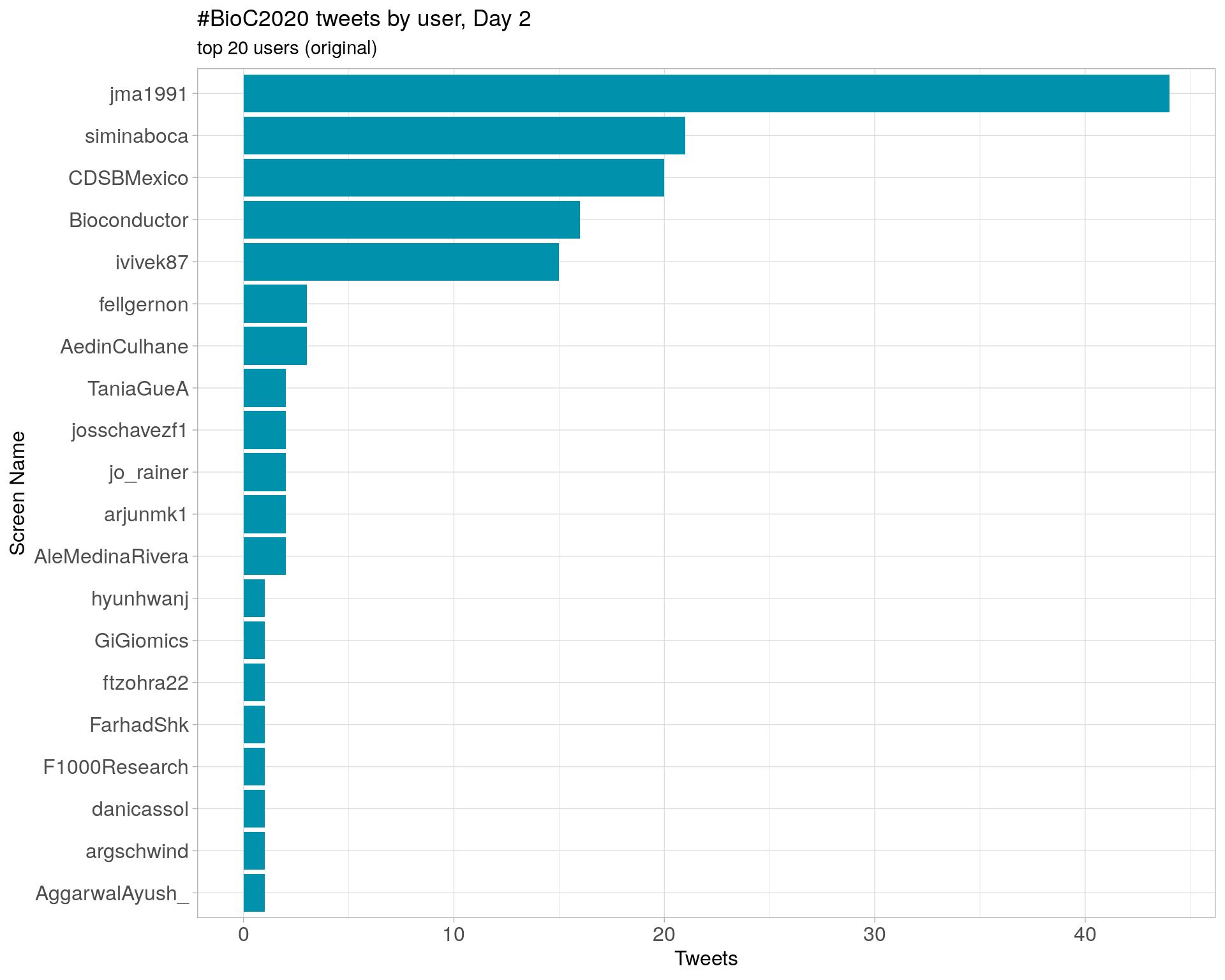
Day 3
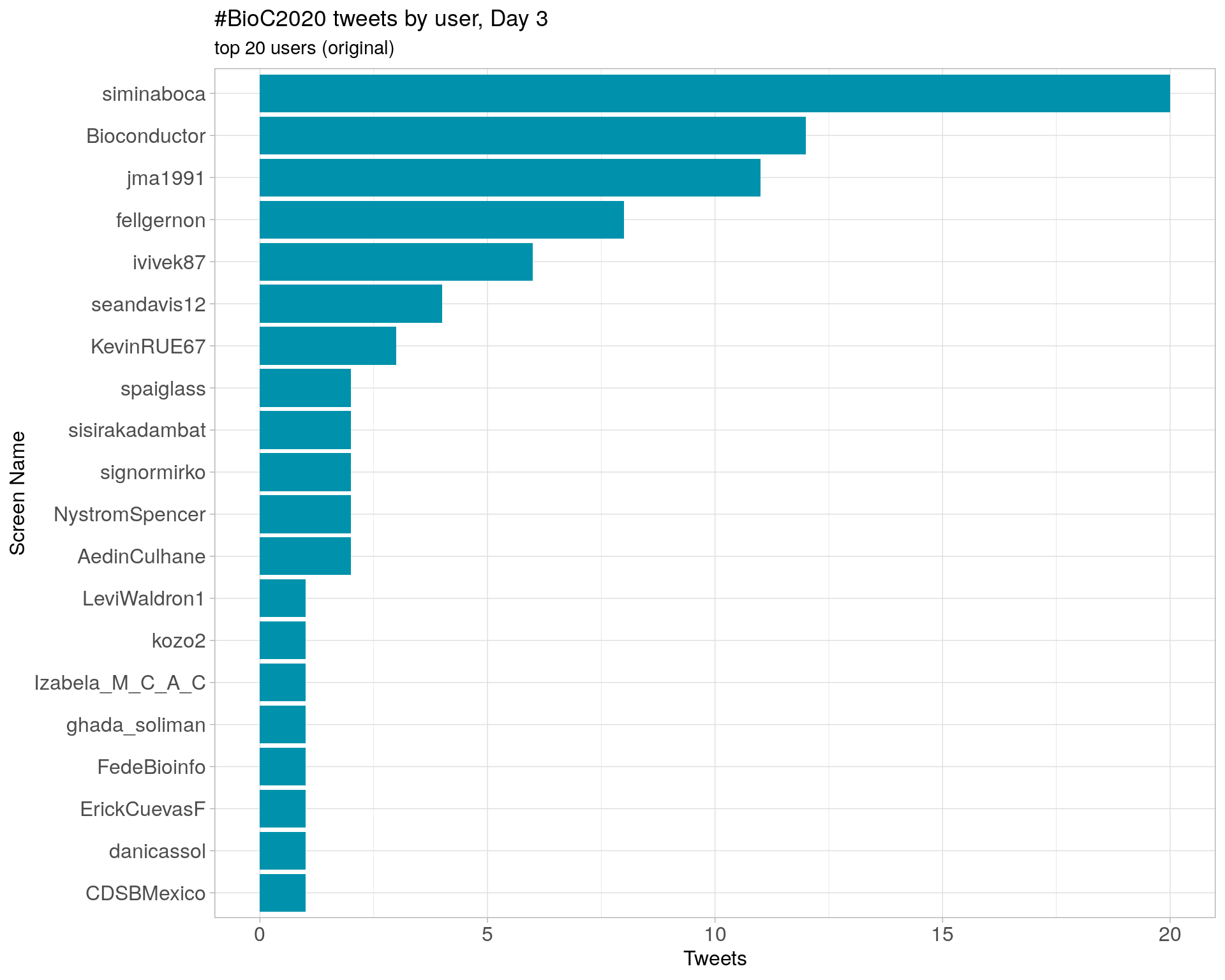
Day 4
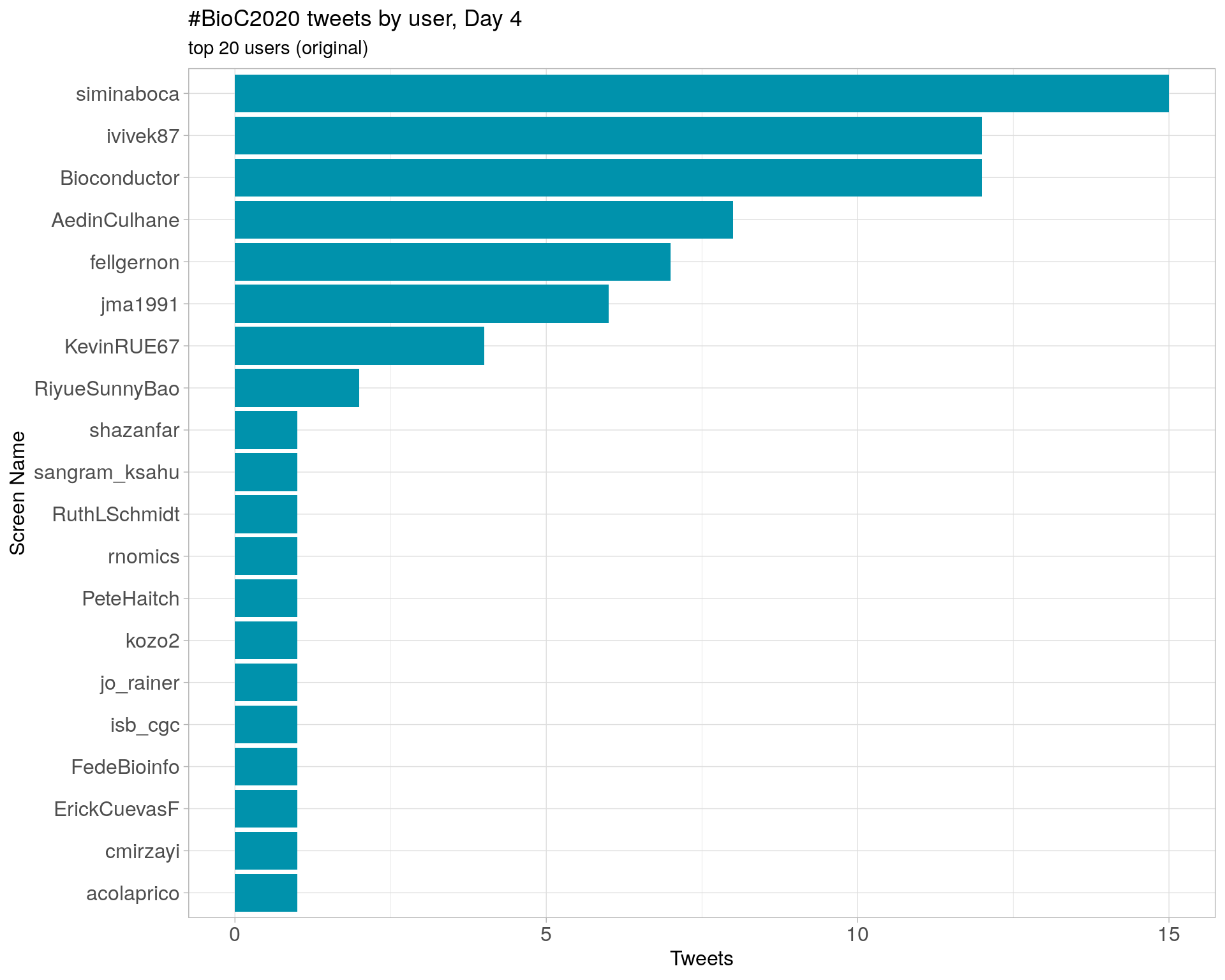
Day 5
Retweets
Day 1
Day 2

Day 3
Day 4
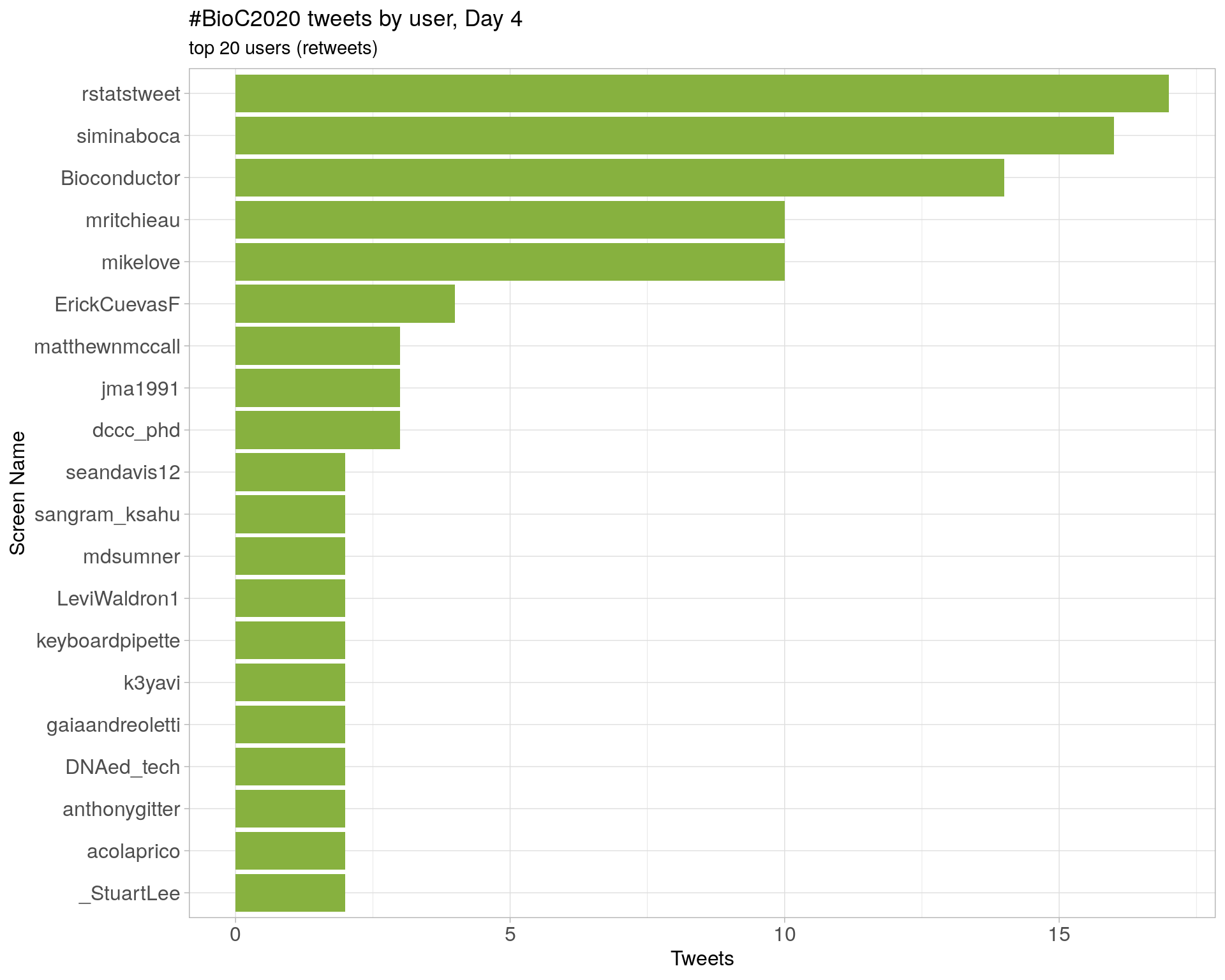
Day 5
4 Sources

5 Networks
5.1 Replies
The “replies network”, composed from users who reply directly to one another, coloured by PageRank.

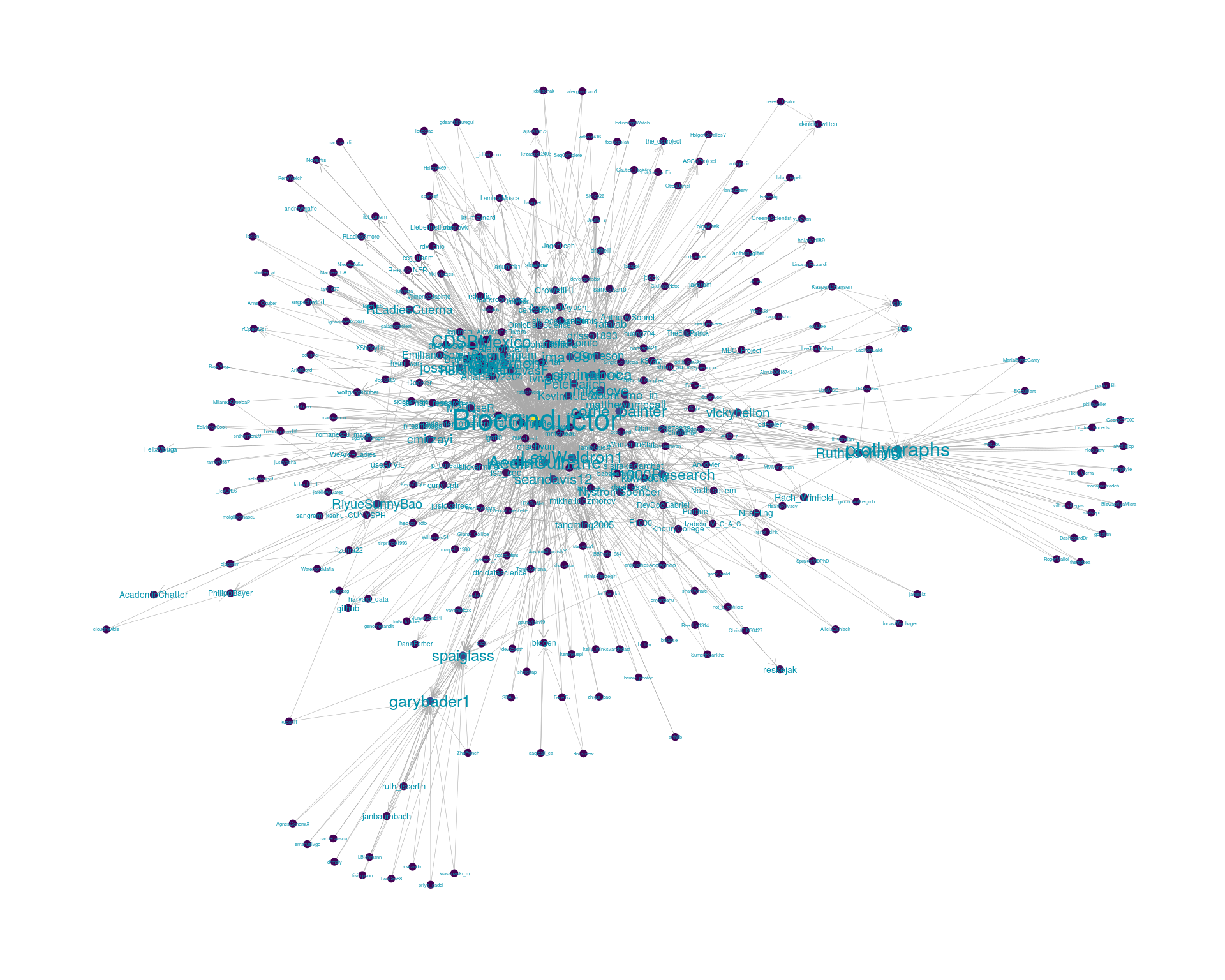
5.2 Mentions
The “mentions network”, where users mention other users in their tweets. Filtered for a k-core of 2. Node colour and size adjusted according to PageRank score.
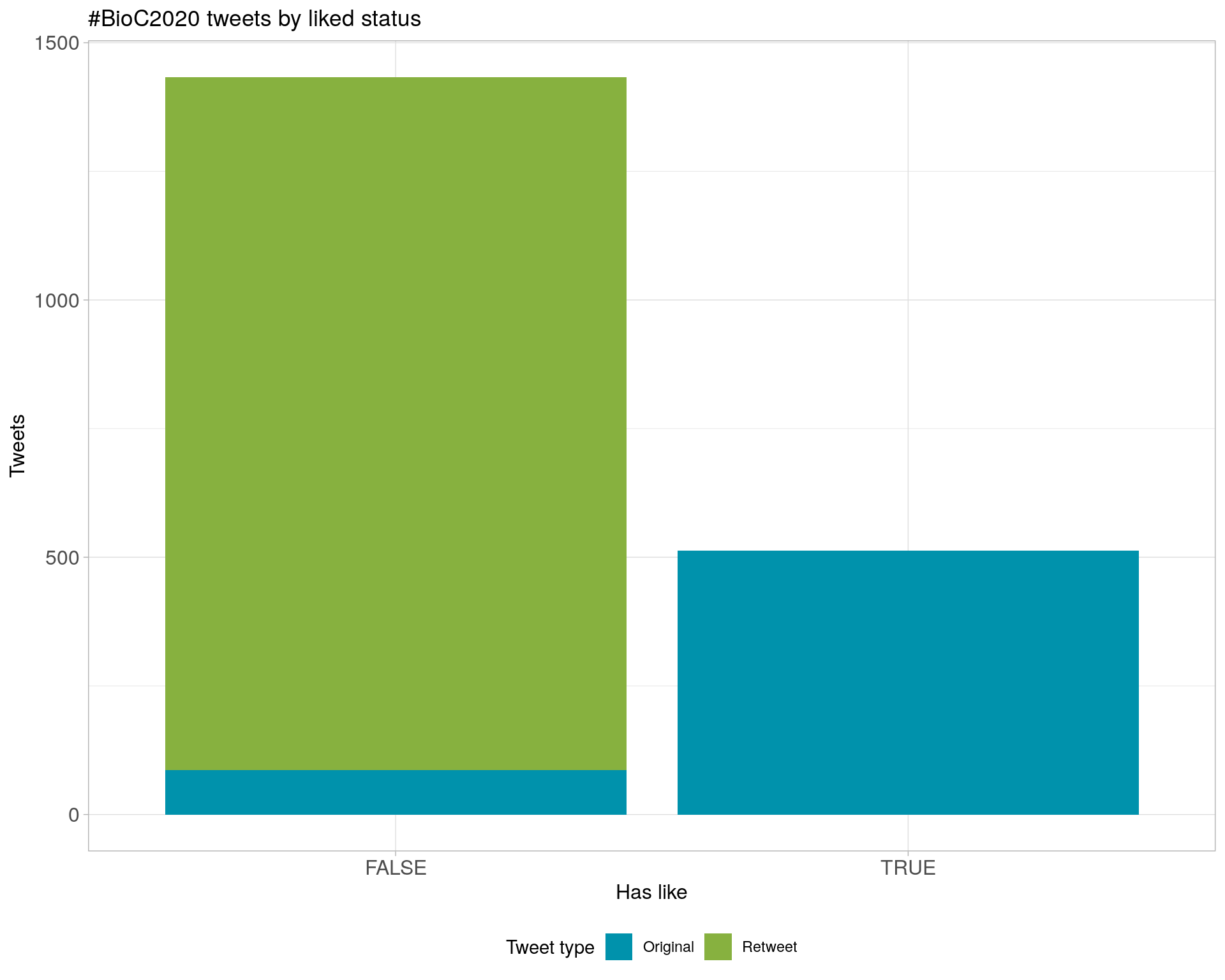
6 Tweet types
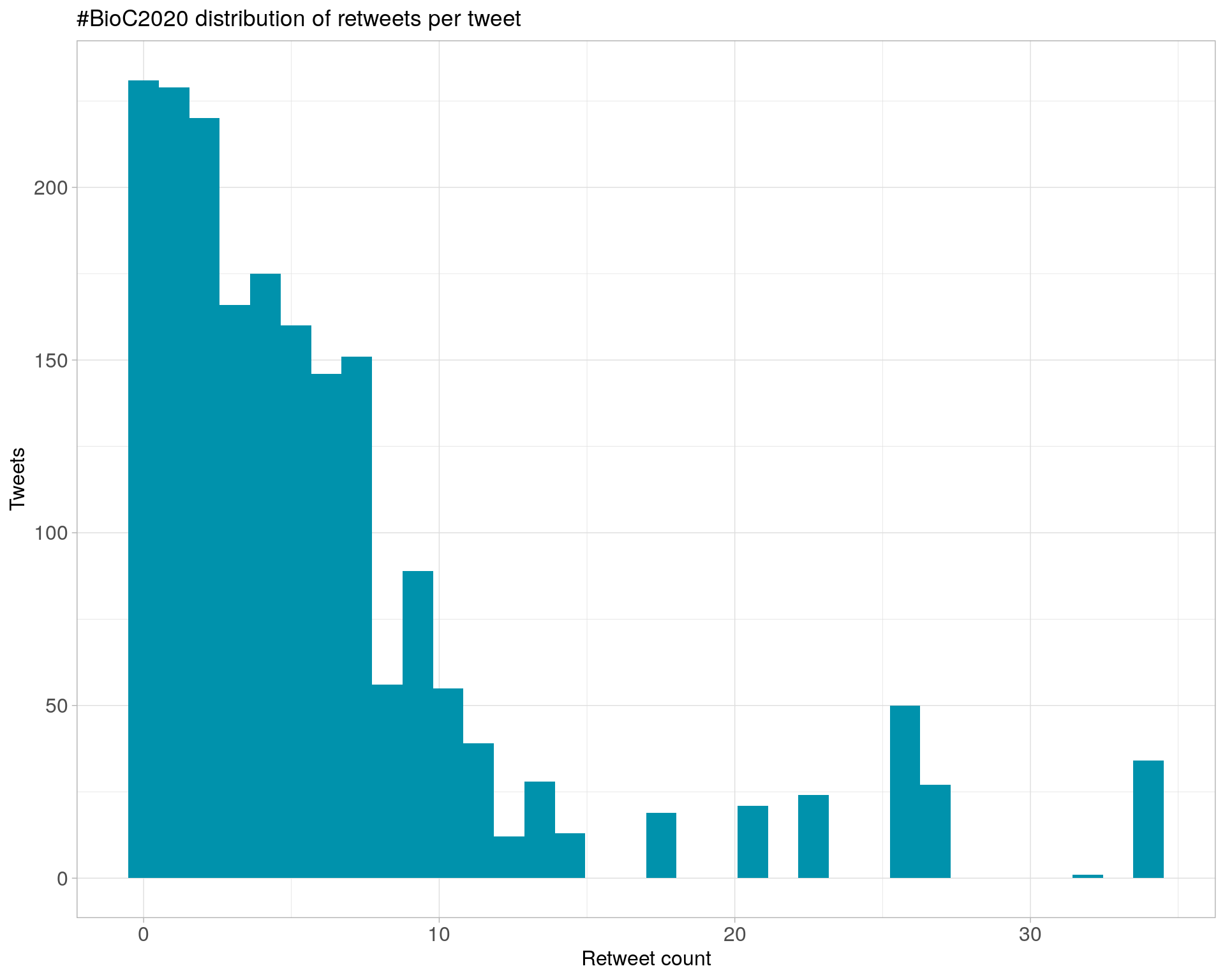
6.1 Retweets
Proportion

Count
Top 10
| screen_name | text | retweet_count |
|---|---|---|
| LeviWaldron1 | Here is the full collection of posters for @Bioconductor #bioc2020. Slides for at least some talks will be posted there too, and a couple are up. https://t.co/QOOuTlvL3Z. This thread lists what’s there now - comments welcome. | 34 |
| RiyueSunnyBao |
In #BioC2020 #BoF session “10 simple rules of #thriving in #bioinformatics #research”. Let’s say NO to - isolated position - unrecognized contribution - neverending projs Let’s say YES to - team environment - well-managed expectation - data-driven research @AcademicChatter |
27 |
| fellgernon |
Tomorrow I’ll be teaching 🎓 a workshop on #recount2 for #BioC2020 It’ll likely be my last #recount2 workshop 😢😇 🌐 https://t.co/navj34rcq4 💻 https://t.co/sDlZfkKMTo 🎚️🌈 https://t.co/Rwy2pPrNJk 🎅🏾🎁 is coming later in 2020 at a @Bioconductor 🪞 near you!☺️ #rstats #RNAseq https://t.co/EhIIwXPvsq |
26 |
| RuthLSchmidt | Excited to be giving a talk today at #bioc2020 about how to build @plotlygraphs dashboards using Dash for R and Dash Bio for #microbiome omics data (like this one below for #metabolomics data). Tune in from 3-4 pm EST. Slides and talk will also be made available after. #dataviz https://t.co/VOU2QzOOEv | 26 |
| PeteHaitch |
Whether or not you’re at #bioc2020, you can use the instructional materials and watch a recording of my workshop on effectively using the DelayedArray framework to analyse large datasets with #rstats Workshop material: https://t.co/dOjICGcgVF Video: https://t.co/a2nLBdRynF |
23 |
| spaiglass | Excited to present our software at #bioc2020 Thursday 3-4pm. Work with @garybader1 netDx: Build a patient classifier that integrates multi ’omic data, uses machine learning to predict clinical outcome. Now in @Bioconductor: https://t.co/uhoMRuTlPd Software paper coming soon! /1 https://t.co/IV38RbE7SI | 21 |
| mikelove |
Congrats to the Bioc community award winners! 🎉 The motivation for these award winners is quite impressive #bioc2020 Helena Crowell @CrowellHL Aaron Lun Lori Shepherd @lshep712 Gordon Smyth |
18 |
| AedinCulhane |
#Bioc2020 are extremely grateful to phenomenal @seandavis12 who is co-ordinating everything to run 22 live workshops He created a @github template for workshop presenters. It uses GitHub Actions to pkgdown websites and creates a docker image of each pkg https://t.co/wo9x7ccTxo |
14 |
| Bioconductor |
Chloe @cmirzayi shows how easily you can make causal diagrams using #daggity 📦 #rstats #BioC2020 https://t.co/8cEElGwANs |
13 |
| josschavezf1 | Join me tomorrow in the poster session A at #BioC2020 I will show you how to use my recent package #erba to study transcriptional regulators from bacteria and archaea 🧬 https://t.co/vu6YbNYgM7 | 13 |
Most retweeted
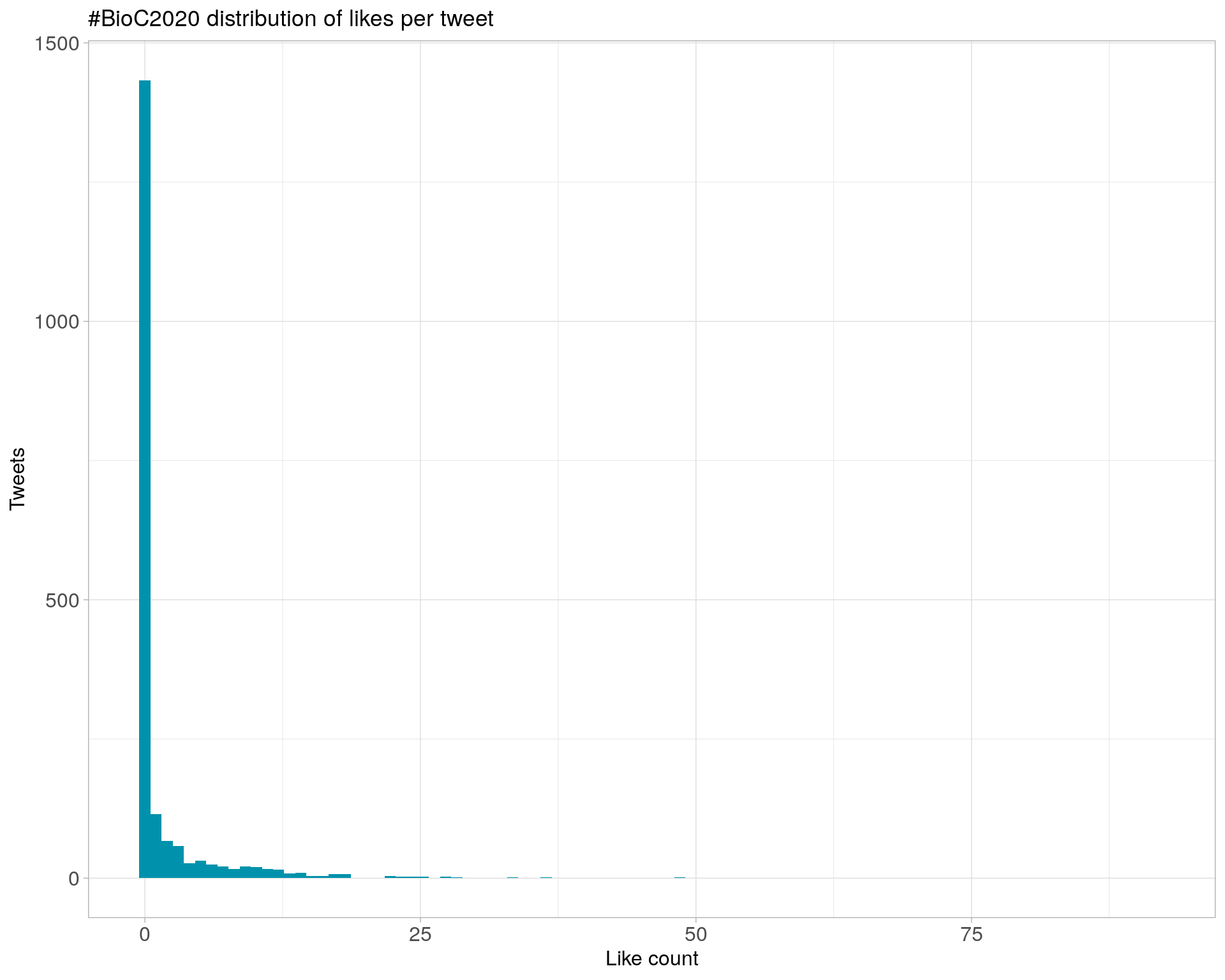
6.2 Likes
Proportion
Count
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| fellgernon |
Tomorrow I’ll be teaching 🎓 a workshop on #recount2 for #BioC2020 It’ll likely be my last #recount2 workshop 😢😇 🌐 https://t.co/navj34rcq4 💻 https://t.co/sDlZfkKMTo 🎚️🌈 https://t.co/Rwy2pPrNJk 🎅🏾🎁 is coming later in 2020 at a @Bioconductor 🪞 near you!☺️ #rstats #RNAseq https://t.co/EhIIwXPvsq |
92 |
| RuthLSchmidt | Excited to be giving a talk today at #bioc2020 about how to build @plotlygraphs dashboards using Dash for R and Dash Bio for #microbiome omics data (like this one below for #metabolomics data). Tune in from 3-4 pm EST. Slides and talk will also be made available after. #dataviz https://t.co/VOU2QzOOEv | 81 |
| RiyueSunnyBao |
In #BioC2020 #BoF session “10 simple rules of #thriving in #bioinformatics #research”. Let’s say NO to - isolated position - unrecognized contribution - neverending projs Let’s say YES to - team environment - well-managed expectation - data-driven research @AcademicChatter |
79 |
| spaiglass | Excited to present our software at #bioc2020 Thursday 3-4pm. Work with @garybader1 netDx: Build a patient classifier that integrates multi ’omic data, uses machine learning to predict clinical outcome. Now in @Bioconductor: https://t.co/uhoMRuTlPd Software paper coming soon! /1 https://t.co/IV38RbE7SI | 77 |
| mikelove |
Congrats to the Bioc community award winners! 🎉 The motivation for these award winners is quite impressive #bioc2020 Helena Crowell @CrowellHL Aaron Lun Lori Shepherd @lshep712 Gordon Smyth |
74 |
| AedinCulhane |
#Bioc2020 are extremely grateful to phenomenal @seandavis12 who is co-ordinating everything to run 22 live workshops He created a @github template for workshop presenters. It uses GitHub Actions to pkgdown websites and creates a docker image of each pkg https://t.co/wo9x7ccTxo |
64 |
| LeviWaldron1 | Here is the full collection of posters for @Bioconductor #bioc2020. Slides for at least some talks will be posted there too, and a couple are up. https://t.co/QOOuTlvL3Z. This thread lists what’s there now - comments welcome. | 63 |
| seandavis12 | With over 600 workshop instances served on a #kubernetes cluster on #GoogleCloud on just the first day of #bioc2020, the impact of @isb_cgc efforts and resources has been a multiplier for the fantastic work done by Bioconductor workshop contributors. https://t.co/VzVYcOi2Ru | 54 |
| tangming2005 | dplyr-based Access to Bioconductor Annotation Resources https://t.co/XoI1yOGNRT #Bioc2020 | 49 |
| PeteHaitch |
Whether or not you’re at #bioc2020, you can use the instructional materials and watch a recording of my workshop on effectively using the DelayedArray framework to analyse large datasets with #rstats Workshop material: https://t.co/dOjICGcgVF Video: https://t.co/a2nLBdRynF |
49 |
Most likes

6.3 Quotes
Proportion

Count

Top 10
| screen_name | text | quote_count |
|---|---|---|
| ivivek87 |
@siminaboca @OmicDataScience @fellgernon Thoroughly enjoyed the last workshop session of Day 1 #BioC2020 https://t.co/P2JIODWf6T https://t.co/J73N6DqMAB |
4 |
| AggarwalAyush_ | Really showing the power and easiness in using #tidyverse for #rnaseq analysis. Amazing workshop by @steman_research and maria doyle. My 1st bioc conference is turning out to be amazing. Lucky that it is virtual this time #BioC2020 #RStats https://t.co/Wgg85D1d2b | 4 |
| Bioconductor |
@WeAreRLadies A @Bioconductor package ;) #BioC2020 help out @romanescu_maria with these questions! We know that many BioC users are #tidyverse 🤩 fans. Some of you even make new packages to bridge both worlds, like @steman_research #rstats https://t.co/56TySdFPJ6 |
4 |
| TaniaGueA |
on #tidybulk: Shut up and take my data 🤓 Thanks for the very nice explanation! #BioC2020 https://t.co/kCKTcTzojI https://t.co/MPDwUYlq4v |
4 |
| ivivek87 |
Very insightful workshop @cmirzayi on causal diagrams. #BioC2020 https://t.co/Qu7SBjYbBJ https://t.co/t5PVd9Ocoy |
3 |
| LeviWaldron1 | Outstanding #bioc2020 workshop today by @cmirzayi of @cunyisph and @CUNYSPH, to a live audience of 140. If you missed it, https://t.co/lhyrHSz5ml https://t.co/wVoNbspX4s | 3 |
| TaniaGueA | I feel enlighted! Looking forward to try out #daggity on my own data and sit and cry once I confirm I did everything wrong the first time! lol🙈but at least now I know! Amazing talk and I’M LOVING THE WORKSHOPS PLATFORM! ❤️ #BioC2020 https://t.co/nlwAGlcHZd | 3 |
| ivivek87 |
Day 2 begins at #bioc2020 😃 with Gene, networks and high throughout Omics. #Bioinformatics #RStats #bioconductor https://t.co/vxlo7deu7S |
3 |
| arjunmk1 | Darn! I missed this one! However, I can watch the recorded talk and all the workshop data is available in one click! thanks, #bioc2020 organizers! https://t.co/Gk7qku54nh | 3 |
| ftzohra22 | Day 2 starting with gene-set analysis workshop #BioC2020 https://t.co/Penu5YPCsV | 3 |
Most quoted
7 Media
Proportion

Top 10
| screen_name | text | favorite_count |
|---|---|---|
| fellgernon |
Tomorrow I’ll be teaching 🎓 a workshop on #recount2 for #BioC2020 It’ll likely be my last #recount2 workshop 😢😇 🌐 https://t.co/navj34rcq4 💻 https://t.co/sDlZfkKMTo 🎚️🌈 https://t.co/Rwy2pPrNJk 🎅🏾🎁 is coming later in 2020 at a @Bioconductor 🪞 near you!☺️ #rstats #RNAseq https://t.co/EhIIwXPvsq |
92 |
| RuthLSchmidt | Excited to be giving a talk today at #bioc2020 about how to build @plotlygraphs dashboards using Dash for R and Dash Bio for #microbiome omics data (like this one below for #metabolomics data). Tune in from 3-4 pm EST. Slides and talk will also be made available after. #dataviz https://t.co/VOU2QzOOEv | 81 |
| spaiglass | Excited to present our software at #bioc2020 Thursday 3-4pm. Work with @garybader1 netDx: Build a patient classifier that integrates multi ’omic data, uses machine learning to predict clinical outcome. Now in @Bioconductor: https://t.co/uhoMRuTlPd Software paper coming soon! /1 https://t.co/IV38RbE7SI | 77 |
| AedinCulhane |
#Bioc2020 are extremely grateful to phenomenal @seandavis12 who is co-ordinating everything to run 22 live workshops He created a @github template for workshop presenters. It uses GitHub Actions to pkgdown websites and creates a docker image of each pkg https://t.co/wo9x7ccTxo |
64 |
| josschavezf1 | Join me tomorrow in the poster session A at #BioC2020 I will show you how to use my recent package #erba to study transcriptional regulators from bacteria and archaea 🧬 https://t.co/vu6YbNYgM7 | 47 |
| AedinCulhane | #bioc2020 is grateful to @stickermule for in-kind sponsorship so participants can get conference stickers. The @Bioconductor community’s expanding collection of package, event & diversity hex stickers are at https://t.co/gpV0EYNpKE Add a sticker with pkg hexSticker::sticker https://t.co/OkQHAluEZV | 41 |
| Bioconductor |
For our final activity today we have Mike Love @mikelove Avi Srivastava @k3yavi on “Importing #alevin scRNA-seq counts into R/Bioconductor” In case you don’t know, Mike is a star 🤩 at the Bioconductor Support Website #BioC2020 #rstats https://t.co/H3DBP0lIAu |
37 |
| Bioconductor |
Thank you everyone for attending #BioC2020!!! 🤗 You are the community & what gives life to the project! 🤩 Levi Waldron @LeviWaldron1 Aedin Culhane @AedinCulhane now give the closing remarks😢 Thank you organizing team! It’s all volunteers & you can volunteer next years too! https://t.co/k6fUxeKHdM |
36 |
| mikelove |
For a #bioc2020 speaker it’s nice to see attendees adding a workshop to their schedules in advance 😁 Talks are recorded and workshop material is online. My and @k3yavi’s vignette on importing alevin scRNA-seq counts with tximeta (work w @nomad421 et al) https://t.co/bVbJJnZcHH https://t.co/htI3Lox2sr |
36 |
| AedinCulhane | #bioc2020 Dan Bunis, Atul Deskpande ask for colorblind friendly figures. Critical for single cell t-SNE/UMAP @humancellatlas Faceting figures, overlaid labels or even just letters (A,B,C) on clusters. Recommend less than 8 colors. dittoSeq sets a new standard for all #rstats https://t.co/xtAeY9kncD | 35 |
7.1 Most liked image

8 Tweet text
8.1 Word cloud
The top 100 words used 3 or more times.

8.2 Bigram graph
Words that were tweeted next to each other at least 3 times.

8.3 Topic modelling
Top 10 words associated with 6 topics identified by LDA.
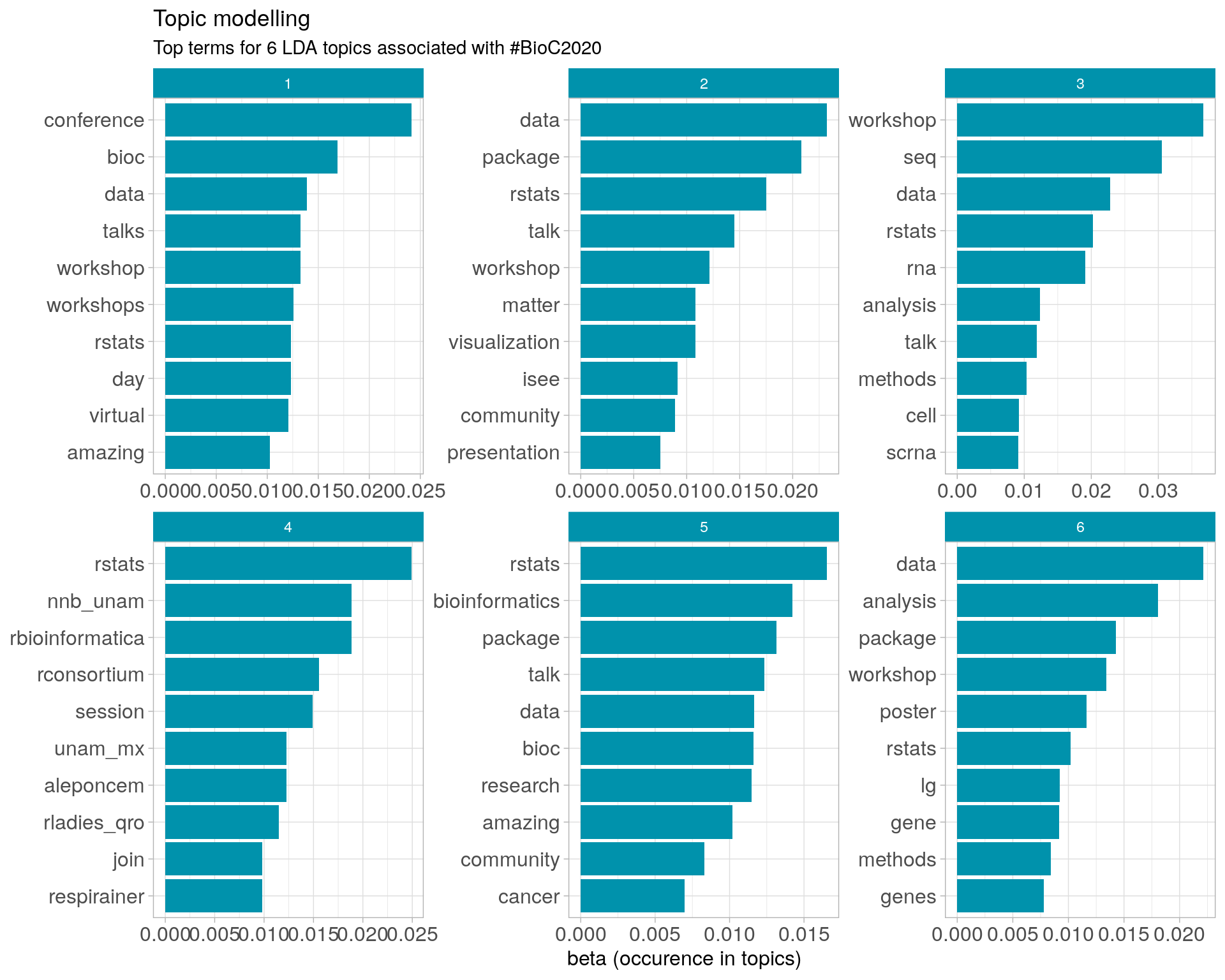
8.3.1 Representative tweets
Most representative tweets for each topic
Topic 1
| screen_name | text | gamma |
|---|---|---|
| Bioconductor |
@niteshturaga @mt_morgan @kaylainter1011 @lshep712 @M2RUseR @Docker @rstudio Learn more details about the BioC @Docker images by @niteshturaga et al (based on #rocker by @rOpenSci members) at Info on rocker: https://t.co/29Yp2JKhSV @noamross @cboettig @eddelbuettel #BioC2020 #rstats (#TakeTwo messed up my from acc the 1st time) https://t.co/cnufYROdx6 |
0.9959096 |
| Bioconductor |
We now continue with X. Shirley Liu @XShirleyLiu to learn about “Computational modeling of protein degradation in tumors” Her research focuses on algorithm dev & integrative mining from big data generated on microarrays, massively parallel sequencing & HT tech #BioC2020 #rstats https://t.co/o9sS0TisZi |
0.9952839 |
| Bioconductor |
Nitesh @niteshturaga Martin @mt_morgan Kayla @kaylainter1011 Lori @lshep712 Marcel @M2RUseR & more at the BioC team are showing how you can use @Docker to simplify your BioC #rstats experience Coming soon: binary installations like the RSPM @rstudio & newer ubuntu😍😎 #BioC2020 https://t.co/DfjxmhGYR1 |
0.9946727 |
| AedinCulhane | #bioc2020 virtual conference is next week. Our virtual platform is at capacity & registration is closed. If you missed out, info on where to view posters and workshop materials are on https://t.co/MofhfF8m8k. Talks will be recorded & videos pushed to youtube after the conference https://t.co/ltlKBOta6z | 0.9944322 |
| F1000Research | We’re thrilled to be supporting #BioC2020! To celebrate, we’re highlighting the awesome @Bioconductor Gateway on @F1000Research, so stay tuned this week for our top picks in #bioinformatics research 😊 And don’t forget to check out the conference posters: https://t.co/4WOQBJ4PIk https://t.co/7s9ZPxO0IB | 0.9944322 |
| RiyueSunnyBao |
#BioC2020 day 1 kicking off!!!!!!!! Lots of super exciting #talks & #workshops! We are also actively #recruiting scientists to join our fight against #cancer @UPMCHillmanCC. Check job board!! You will find me on forum, channel, here and there … open to coffee chat! :) https://t.co/9dI4h9BRi4 |
0.9944322 |
| Bioconductor |
Final keynote; Aaron Lun talks about “Making the infrastructure sausage: tales of Bioconductor package development” Aaron is 1 of the inaugural 4 Bioconductor Community award winners 🎖️, promises to have 20 min for questions & is a 📦dev all🌟 #BioC2020 https://t.co/6BeCQCjEkF https://t.co/aXQ3NFLWrU |
0.9941689 |
| AedinCulhane | Great tips for #bioc2020 talk. My 3 tips 1) Excite me, What motivated you? Why should I listen? 2) Tell a story, what you did & what it means 3) Easy-2-read slides, Labels esp x,y axis in big font. Folks will read or listen not both together. Your 3 tips? https://t.co/3IbpqHMSxs | 0.9938795 |
| siminaboca | Ben Haibe-Kains @bhaibeka now introducing the PharmacoGx package https://t.co/2mtYcIotDg for drug sensitivity data (eg NCI60) and the ToxicoGx package https://t.co/RKEEsPWk9Z which has healthy cell data (don’t want drugs to be toxic to good cells.) #bioc2020 | 0.9938795 |
| mikelove |
Congrats to the Bioc community award winners! 🎉 The motivation for these award winners is quite impressive #bioc2020 Helena Crowell @CrowellHL Aaron Lun Lori Shepherd @lshep712 Gordon Smyth |
0.9935598 |
| AedinCulhane | #bioc2020 is grateful to @isb_cgc for cloud services to run live hands-on workshops during #bioc2020 It wouldn’t be possible without their cloud services and the expertise of @seandavis12. Check out https://t.co/mlB11qXPpN to run BigQuery & DataExplorer on genomics data, TCGA https://t.co/UWxDaANiBC | 0.9935598 |
Topic 2
| screen_name | text | gamma |
|---|---|---|
| AleMedinaRivera |
Hace dos años iniciaron los cursos de la @CDSBMexico gracias al entusiasmo de @areyesq @fellgernon y Heladia Salgado. Esta semana varios miembros de @CDSBMexico participan en #BioC2020. Se ve el resultado del esfuerzo de todos los miembros 🤗 @RBioinformatica @nnb_unam https://t.co/OjGiH76wxv |
0.9963888 |
| Bioconductor |
Julie Zhu & Jianhong Ou are starting their workshop on “CRISPRseek for design target-specific gRNAs for the CRISPR genome editing system including base editor and prime editor” #BioC2020 #rstats #CRISPR #GenomeEditing https://t.co/sBe92FNHqp |
0.9952839 |
| AedinCulhane | #bioc2020 are grateful MAZE Therapeutics for their sponsorships. MAZE have job openings including a Computational Biologist/Senior Computational Biologist position based in Maze Therapeutics (South San Francisco, CA). See https://t.co/xgDbR3biM9 Please RT https://t.co/2EWFwUkBVc | 0.9948933 |
| Bioconductor |
For our 1st talk today: Kylie Bemis @kuwisdelu on “Out-of-memory computing with matter” Beyond #bioinformatics Kylie is active in outreach to the Native American & LGBTQ communities, an enrolled member of the Zuni tribe + poet & writer #BioC2020 #rstats https://t.co/Z5rBZEgFGX |
0.9946727 |
| siminaboca | Excited about Kylie Bemis @kuwisdelu’s talk today about the matter package https://t.co/7NccffPm0e. KB’s package Cardinal https://t.co/tpmHxOJJQl won the John M. Chambers Statistical Software Award by the American Statistical Association. #bioc2020 https://t.co/6vkw9Vdbg3 | 0.9944322 |
| siminaboca | The matter package uses the concept of atoms “atom = a contiguous sequence of data elements.” Atoms can come from different files. A matter object builds data from atoms. (Matter = many atoms. Glad I got this!) Manuscript available at https://t.co/23mSRmBSq8 #bioc2020 | 0.9941689 |
| siminaboca | At Broad, CP helped launch the “Metastatic Breast Cancer Project” https://t.co/dEb83Wp4zD @MBC_Project. This is a patient-centered study that makes it easy to sign up, vs. having it be driven top-down, allowing a very large number of individuals to participate. #bioc2020 | 0.9941689 |
| Bioconductor |
Charlotte @CSoneson Federico @FedeBioinfo Kevin @KevinRUE67 Aaron Lun are now starting their workshop on #iSEE “nteractive visualization of SummarizedExperiment objects with iSEE” It won one of the awards in the first @rstudio #shiny contest 🌟🏆 #BioC2020 #rstats https://t.co/qE6ikppXd1 |
0.9938795 |
| RuthLSchmidt | Excited to be giving a talk today at #bioc2020 about how to build @plotlygraphs dashboards using Dash for R and Dash Bio for #microbiome omics data (like this one below for #metabolomics data). Tune in from 3-4 pm EST. Slides and talk will also be made available after. #dataviz https://t.co/VOU2QzOOEv | 0.9938795 |
| siminaboca | Responding to question on balancing data availability and privacy, CP said they use standard approaching like binning years and not releasing germline data. Also essential to always continue dialogue with patients/participants. It’s not done once data are collected! #bioc2020 | 0.9938795 |
Topic 3
| screen_name | text | gamma |
|---|---|---|
| Bioconductor |
Thanks to @seandavis12 & workshop authors you can run the #BioC2020 workshop code on the cloud, your laptop or view 🖥️ ☁️ https://t.co/TyQWGtdDat (7GB RAM, 1.5 CPUs, max 3 hrs) 🐬https://t.co/j1vVMBcmAY (Docker, same #rstats env) 🌍 or view online thx to #pkgdown @github actions https://t.co/hvgoueLSU3 |
0.9952839 |
| Bioconductor |
We continue with Fei Chen on “Slide-seq: a platform for understanding cellular circuits in tissue” Chen’s lab utilizes ExM as a platform for in situ transcriptomics & epigenomics, while pioneering novel molecular & microscopy tools #BioC2020 #rstats https://t.co/juIhCDNbgQ |
0.9950964 |
| siminaboca | RI urges caution with RNAseq results that claim discovery of new cell types. One issue is that scRNA-seq “produces more 0s than expected and.. this bias is greater for lower expressed genes.” This can vary cell-to-cell and confound results. #bioc2020 | 0.9948933 |
| mikelove |
For a #bioc2020 speaker it’s nice to see attendees adding a workshop to their schedules in advance 😁 Talks are recorded and workshop material is online. My and @k3yavi’s vignette on importing alevin scRNA-seq counts with tximeta (work w @nomad421 et al) https://t.co/bVbJJnZcHH https://t.co/htI3Lox2sr |
0.9948933 |
| Bioconductor |
Our next contributed talks session is starting with Lambda Moses @LambdaMoses Ellis Patrick @TheEllisPatrick Dario Righelli @drighelli Lukas M Weber @lmwebr Learn about #spatialLIBD, #VisiumExperiment, #smFISH data methods, and spicyR #BioC2020 #rstats #spatial #Visium https://t.co/U399TnwUDH |
0.9948933 |
| Bioconductor |
Next up we have Leonardo Collado Torres @fellgernon for our 4th workshop: “Human RNA-seq data from #recount2 and related packages” It’s his 3rd BioC recount2 workshop (2017, 19, 20) Leo will also present a poster & BoF at #BioC2020 #rstats #RNAseq https://t.co/BsY8z1LfOB |
0.9946727 |
| siminaboca | After the success with #mbc, CP wanted to start a similar project with angiosarcoma. Angiosarcoma is much rarer than breast cancer (300 dx a year), has a poor prognosis, patients are scattered. Got support from the angiosarcoma community, as well as from #mbc community! #bioc2020 https://t.co/hQ1bbOwokH | 0.9944322 |
| siminaboca | 2nd workshop of the day! Chloe Mirzayi @cmirzayi introduces epidemiology for bioinformaticians! Detailed explanations on confounders, mediators, colliders. Selection bias (including loss to follow) can be a form of collider bias. #bioc2020 | 0.9941689 |
| Bioconductor |
Our 6th and last #BioC2020 workshop for today is by Stefano Mangiola @steman_research and Maria Doyle on “A tidy transcriptomics introduction to RNA-Seq analyses” Join us tomorrow for more great content by our community =) #BioC2020 #rstats #RNAseq https://t.co/qVq7Zl9jaK |
0.9941689 |
| Bioconductor |
Now Aedin Culhane @AedinCulhane and Lauren Hsu are teaching their “An introduction to matrix factorization and principal component analysis in R” #BioC2020 workshop Learn about the principles behind PCA + SVD and how it can be applied to #scData #rstats https://t.co/eNxClcAs9h |
0.9941689 |
Topic 4
| screen_name | text | gamma |
|---|---|---|
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar @EmilianoSotel10 @josschavezf1 @RLadiesCuerna @VjimenezJacinto @ibt_unam @rstudio @areyesq @wolfgangkhuber @rafalab @Novartis Leonardo Collado Torres @fellgernon is a co-founder & CDSB member + the 2nd main #CDSB2020 instructor He is a BioC vet, part of the @Bioconductor Community Advisory Board, has 3 #BioC2020 presentations tomorrow: 🚕, poster, BoF on CDSB #BioC2020 #rstats https://t.co/HQWffxbylX |
0.9963888 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar @EmilianoSotel10 @josschavezf1 @RLadiesCuerna @VjimenezJacinto @ibt_unam @rstudio Leticia Vega Alvarado is the 3rd/3 @RLadiesCuerna co-founder & co-instructor with Vero for the intro to R & @rstudio workshop Lety joined CDSB this week also and like Vero is attending BioC for the first time https://t.co/dJOx17EMB4 https://t.co/gvTlhcFZo9 #BioC2020 #rstats https://t.co/jA76Sm9gLO |
0.9962798 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 Carmina Barberena @BarjonCar is a CDSB alumni & one of our first @Bioconductor package new developers thx to #regutools More on this project at the Birds of a Feather session tomorrow Wednesday https://t.co/Tnqu5PBUUI Carmina recently finished her undergrad💯 #BioC2020 #rstats https://t.co/tZB8tJKg28 |
0.9959096 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar @EmilianoSotel10 @josschavezf1 @RLadiesCuerna Veronica Jimenez Jacinto @VjimenezJacinto joined us this week & is a @RLadiesCuerna co-founder Vero taught many of us at @lcgunam analyzes data at @ibt_unam + teaches the intro to R & @rstudio @RBioinformatica @nnb_unam summer workshop #BioC2020 #rstats https://t.co/hHhTc1A3BA |
0.9959096 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar @EmilianoSotel10 Our 3rd @Bioconductor new developer is Joselyn Chavez @josschavezf1 & #regutools maintainer Co-founded @RLadiesCuerna after #CDSB2018 & 19, won scholarships for #BioC2019 #rstudioconf 2020, is #CDSB2020 main instructor & board member🤩 #BioC2020 #rstats https://t.co/U4PALb1kag |
0.9956190 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca Yalbi Balderas @yalbi_ibm is a prof at @RespiraINER working on lung diseases including COVID-19 Yalbi is a great person to collaborate with! If you have a grant proposal idea, get in touch with her. Her local exp will be invaluable 💯 #BioC2020 #rstats https://t.co/0OLYkhnhBY |
0.9954576 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar @EmilianoSotel10 @josschavezf1 @RLadiesCuerna @VjimenezJacinto @ibt_unam @rstudio Alejandro Reyes @areyesq is a co-founder & CDSB board member You might know him for his #DEXSeq work with @wolfgangkhuber, but he’s also behind many other packages wth @rafalab & more including #regutools He recently joined @Novartis #BioC2020 #rstats https://t.co/aGUfZeKqdM |
0.9954576 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio Marcel Ramos @M2RUseR recently joined @CDSBMexico after volunteering to teach part of #CDSB2020 coming up soon Marcel is part of the @Bioconductor core team, works with @LeviWaldron1 & co in many key 📦 including #MultiAssayExperiment #BioC2020 #rstats https://t.co/fpmzF9NFQJ |
0.9952839 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam @rdv_chio @M2RUseR @Bioconductor @LeviWaldron1 @BarjonCar Along with Carmina, Jesus Emiliano Sotelo Fonseca @EmilianoSotel10 worked on #regutools and is a @Bioconductor package developer 🙌🏽 Emi worked on #praiseMX https://t.co/17t6Fq1n79 during #CDSB2019 It’s a hilarious 📦, check it out! #BioC2020 #rstats https://t.co/lF8rygtgHJ |
0.9952839 |
| CDSBMexico |
@RConsortium @RBioinformatica @nnb_unam @ErickCuevasF @UNAM_MX @Aleponcem @TeresaOM @AnaBetty2304 @RLadies_Qro @markrobinsonca @yalbi_ibm @RespiraINER @lcgunam @ccg_unam Rocío Domínguez Vidaña @rdv_chio is a Cancer Bioinformatician & Data Scientist who started her career at @lcgunam Rocio will be a student at #CDSB2020 next week #BioC2020 #rstats https://t.co/4kEQ5KVnwH |
0.9950964 |
Topic 5
| screen_name | text | gamma |
|---|---|---|
| AedinCulhane | #bioc2020 Dan Bunis, Atul Deskpande ask for colorblind friendly figures. Critical for single cell t-SNE/UMAP @humancellatlas Faceting figures, overlaid labels or even just letters (A,B,C) on clusters. Recommend less than 8 colors. dittoSeq sets a new standard for all #rstats https://t.co/xtAeY9kncD | 0.9952839 |
| siminaboca | MS imaging (MSI) leads to complicated data structure (“data cube”). This led to KB developing Cardinal (w/ PhD adviser @olgavitek, who is curating @WomenInStat this week!), as at the time there were no packages specifically for MS imaging. https://t.co/1dmRKsAQP1 #bioc2020 | 0.9948933 |
| Bioconductor |
You can now choose: “Birds of a Feather: Reproducible environments for integrated computational workflows” with @KevinRUE67 & Charlie George or Talk with the BioC Core Team with @lshep712 & Hervé Pagès on build system, single 📦 builder tracker for new submissions #BioC2020 https://t.co/URYsxuDTFf |
0.9948933 |
| siminaboca | This was the inspiration to create the matter package, using the imzML format as input. imzML = open-source format for interchange of MS imaging experiments, combination of XML file (meta-data) and binary file (spectral data). #bioc2020 | 0.9946727 |
| Bioconductor |
Next, Corrie Painter @corrie_painter on “Count Me In, Partnering with patients to accelerate the pace of cancer research” She has combined her cancer advocacy & scientific bkg to engage with patients in order to build & carry out patient-partnered genomic ⚗️🔎 #BioC2020 #rstats https://t.co/9RTl7uRxzl |
0.9946727 |
| Bioconductor |
Next you have 2 options (though recordings will be available later): Birds of a Feather: Ten simple rules for thriving in bioinformatics research or Talk with the BioC Core Team: open discussion on BiocSet, annotation pipelines for release, package creation/review #BioC2020 https://t.co/txtF8jVPaB |
0.9946727 |
| spaiglass | Excited to present our software at #bioc2020 Thursday 3-4pm. Work with @garybader1 netDx: Build a patient classifier that integrates multi ’omic data, uses machine learning to predict clinical outcome. Now in @Bioconductor: https://t.co/uhoMRuTlPd Software paper coming soon! /1 https://t.co/IV38RbE7SI | 0.9944322 |
| Bioconductor |
Our last contributed talks are by Charlotte Soneson @CSoneson Davide Risso @drisso1893 Anthony Sonrel @AnthonySonrel Stephanie Hicks @stephaniehicks Batch, #scRNAseq, #pipeComp, bench pressing 🏋️♀️🏽🏋️♂️🏽 Includes 2 Technical Advisory Board members 💯 #BioC2020 #rstats #diversity https://t.co/0wOHIXeago |
0.9944322 |
| Bioconductor |
Next up is Ting Huang @huang704 on “MSstatsTMT: Statistical detection of differentially abundant proteins in experiments with isobaric labeling and multiple mixtures” Ting is the second speaker in the session (sorry about the mistake earlier) #BioC2020 #RStats https://t.co/TdRRIPxs2G |
0.9941689 |
| chaos35002422 |
Essential Cell Biology (Day 1 p5) ・Cells are the fundamental units of life ・and diverse in their chemical requirements and activities ・ different from the outside but similar inside ・viruses are chemical zombies #100DaysOfBio #Bioinformatics #Biology #BioC2020 |
0.9941689 |
Topic 6
| screen_name | text | gamma |
|---|---|---|
| PeteHaitch | Ah, there’s nothing like recording oneself to dial up the self-loathing 🥰. Really makes you question your basic skills, e.g. forming a coherent sentence, not to mention your tech skills, e.g. teaching #rstats. PS come to my workshop at #bioc2020 I’ll try not to sound too Strayan https://t.co/FP5XU4GwQj | 0.9948933 |
| Bioconductor |
Our second session of contributed talks is getting started with Daniel Bunis @DanBunis Will Townes @sandakano Lauren Hsu https://t.co/DzYlOsvK0V and Koen Van den Berge @koenvdberge_Be Learn about #dittoSeq, #tradeSeq, scRNAseq dim reduction, & #corral #BioC2020 #rstats https://t.co/PEW3OWNITZ |
0.9948933 |
| ivivek87 |
Day 5 at #bioc2020 starts with “Multi-Domain Data Integration: From Observations to Mechanistic Insights” Very interesting talk using #NeuralNetworks primarily Autoencoder (AE)across data from varied assay types. Lineage tracing using AE in #SingleCell . https://t.co/BoNvt66Mba |
0.9946727 |
| Bioconductor |
For our last level 500 workshop, Kelly Street @justokstreet Koen Van den Berge @koenvdberge_Be and Hector Roux de Bezieux @hector_rdb present “Trajectory inference across conditions: differential expression and differential progression” Join the fun! 💯 #BioC2020 #rstats https://t.co/gZ1fYAJRyd |
0.9946727 |
| Bioconductor |
Hena R Ramay @henaramay Jayaram Kancherla @jayaram Shian Su @shian_su Emma Jablonski are now starting the 5th contributed talks session #RENKU, @nanopore, #FemMicro16S and #epivizrChart are the stars of the show now =) Come learn about them! #BioC2020 #rstats https://t.co/RMu451KgiF |
0.9944322 |
| mikelove |
@Bioconductor @robertclab @jma1991 @NystromSpencer @odemler Only fair given how well James @jma1991 has covered the conference I’ll post his ⚡️ talk #BioC2020 “The Daim software package incorporates methods for pre-processing, normalisation, and peak calling of DamID-seq data” 📦 - https://t.co/fQo0cfVrPv 📜 - https://t.co/RRPQy1YRUR https://t.co/fRXb9Mgf5F |
0.9944322 |
| NystromSpencer |
I’ve been so focused on the cool talks at #BioC2020 I forgot to tweet about my poster! I built an R package which interfaces with the MEME Suite for motif analysis using native R/Bioconductor data structures. Come chat at noon today, or shoot me a DM. https://t.co/AIAqByB7PK |
0.9941689 |
| fellgernon |
@KevinRUE67 @AedinCulhane @Bioconductor I recommend diving this task in 2:
|
0.9941689 |
| Bioconductor |
Now we have Simina Boca @siminaboca on “Using the swfdr package to estimate false discovery rates conditional on covariates” Simina writes excellent twitter summaries, so follow Simina on Twitter for more 📙🔖 #BioC2020 #Rstats https://t.co/zjNBZ8WO76 |
0.9938795 |
| tangming2005 | genes(txdb) can be a problem. e.g. a lncRNA can span the centromere. #Bioc2020 How to define a gene is a problem. I was checking where each gene is located in which cytoband and found some genes span multiple cytobands using genes(txdb) and I found that as well. | 0.9938795 |
9 Software
Software mentioned in Tweets with links to GitHub, BitBucket, Bioconductor or CRAN.
Session info
## R Under development (unstable) (2021-02-18 r80027)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.1 LTS
##
## Matrix products: default
## BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices datasets utils methods base
##
## other attached packages:
## [1] fs_1.5.0 here_1.0.1 kableExtra_1.3.1
## [4] knitr_1.30 magick_2.5.2 webshot_0.5.2
## [7] viridis_0.5.1 viridisLite_0.3.0 wordcloud_2.6
## [10] RColorBrewer_1.1-2 ggraph_2.0.4 ggrepel_0.8.2
## [13] ggplot2_3.3.2 topicmodels_0.2-11 tidytext_0.2.6
## [16] igraph_1.2.6 stringr_1.4.0 purrr_0.3.4
## [19] forcats_0.5.0 lubridate_1.7.9.2 tidyr_1.1.2
## [22] dplyr_1.0.2 rtweet_0.7.0 BiocManager_1.30.10
##
## loaded via a namespace (and not attached):
## [1] httr_1.4.2 tidygraph_1.2.0 jsonlite_1.7.2 assertthat_0.2.1
## [5] askpass_1.1 highr_0.8 stats4_4.1.0 renv_0.12.3
## [9] yaml_2.2.1 slam_0.1-48 pillar_1.4.7 lattice_0.20-41
## [13] glue_1.4.2 digest_0.6.27 polyclip_1.10-0 rvest_0.3.6
## [17] colorspace_2.0-0 plyr_1.8.6 htmltools_0.5.0 Matrix_1.2-18
## [21] tm_0.7-8 pkgconfig_2.0.3 scales_1.1.1 processx_3.4.5
## [25] tweenr_1.0.1 ggforce_0.3.2 tibble_3.0.4 openssl_1.4.3
## [29] generics_0.1.0 farver_2.0.3 ellipsis_0.3.1 withr_2.3.0
## [33] cli_2.2.0 NLP_0.2-1 magrittr_2.0.1 crayon_1.4.1
## [37] ps_1.5.0 evaluate_0.14 tokenizers_0.2.1 janeaustenr_0.1.5
## [41] fansi_0.4.1 SnowballC_0.7.0 MASS_7.3-53 xml2_1.3.2
## [45] tools_4.1.0 lifecycle_0.2.0 munsell_0.5.0 callr_3.5.1
## [49] compiler_4.1.0 rlang_0.4.9 grid_4.1.0 rstudioapi_0.13
## [53] labeling_0.4.2 rmarkdown_2.6 gtable_0.3.0 curl_4.3
## [57] reshape2_1.4.4 graphlayouts_0.7.1 R6_2.5.0 gridExtra_2.3
## [61] utf8_1.1.4 rprojroot_2.0.2 modeltools_0.2-23 stringi_1.5.3
## [65] parallel_4.1.0 Rcpp_1.0.5 vctrs_0.3.5 tidyselect_1.1.0
## [69] xfun_0.19