BioC2019
Bioconductor 2019 conference
2021-02-21 23:30:16
Parameters
| Parameter | Value |
|---|---|
| hashtag | #BioC2019 |
| start_day | 2019-06-24 |
| end_day | 2019-06-27 |
| timezone | America/New_York |
| theme | theme_light |
| accent | #0092ac |
| accent2 | #87b13f |
| kcore | 2 |
| topics_k | 6 |
| bigram_filter | 3 |
| fixed | TRUE |
| seed | 1 |
1 Introduction
An analysis of tweets from the #BioC2019 hashtag for the Bioconductor 2019 conference, 24-27 June 2019 online (originally planned in Boston, USA).
A total of 1563 tweets from 357 users were collected using the rtweet R package.
The latest tweet was recorded at 2019-06-28 14:27:11 (Etc/UTC).
2 Timeline
2.1 Tweets by day
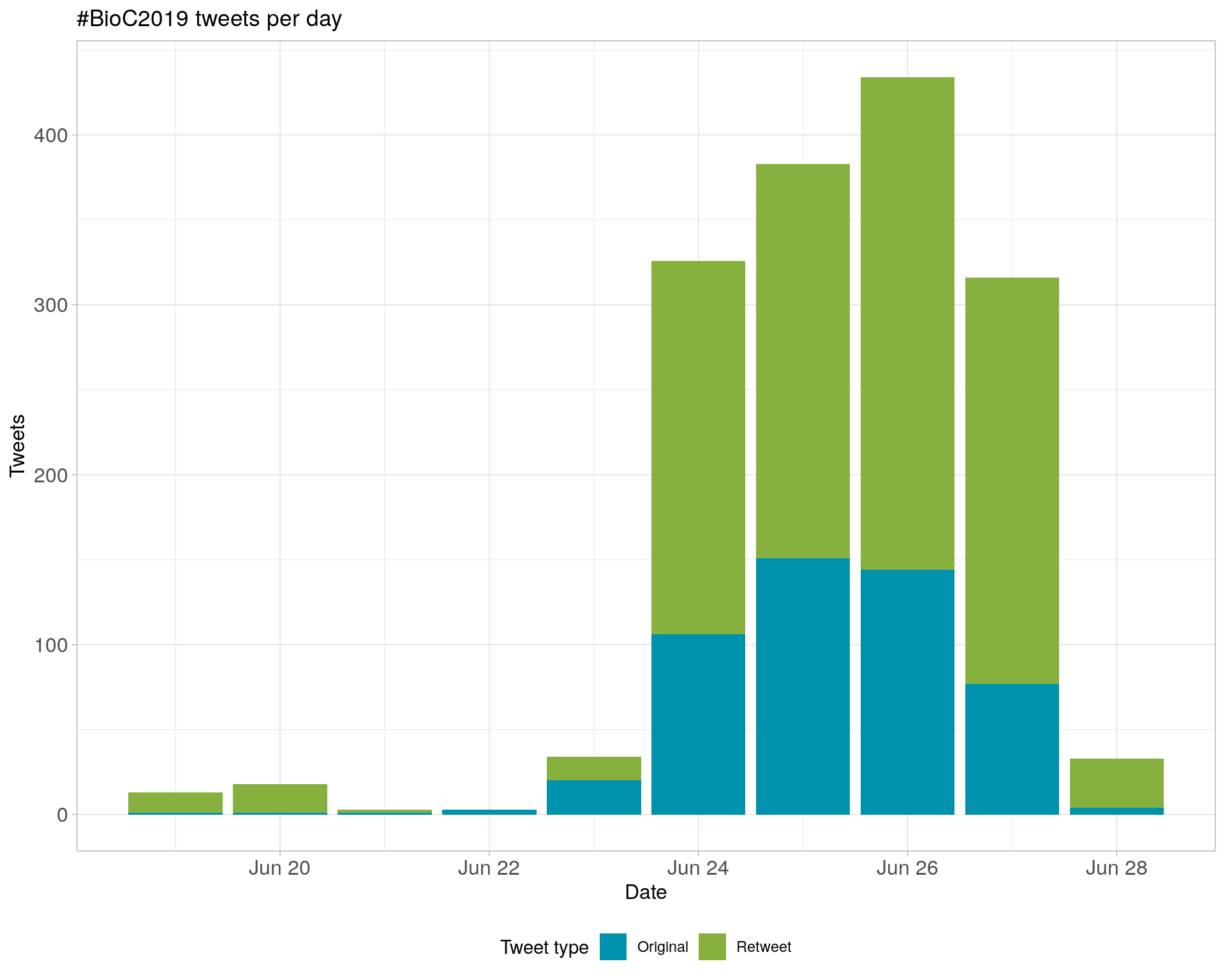
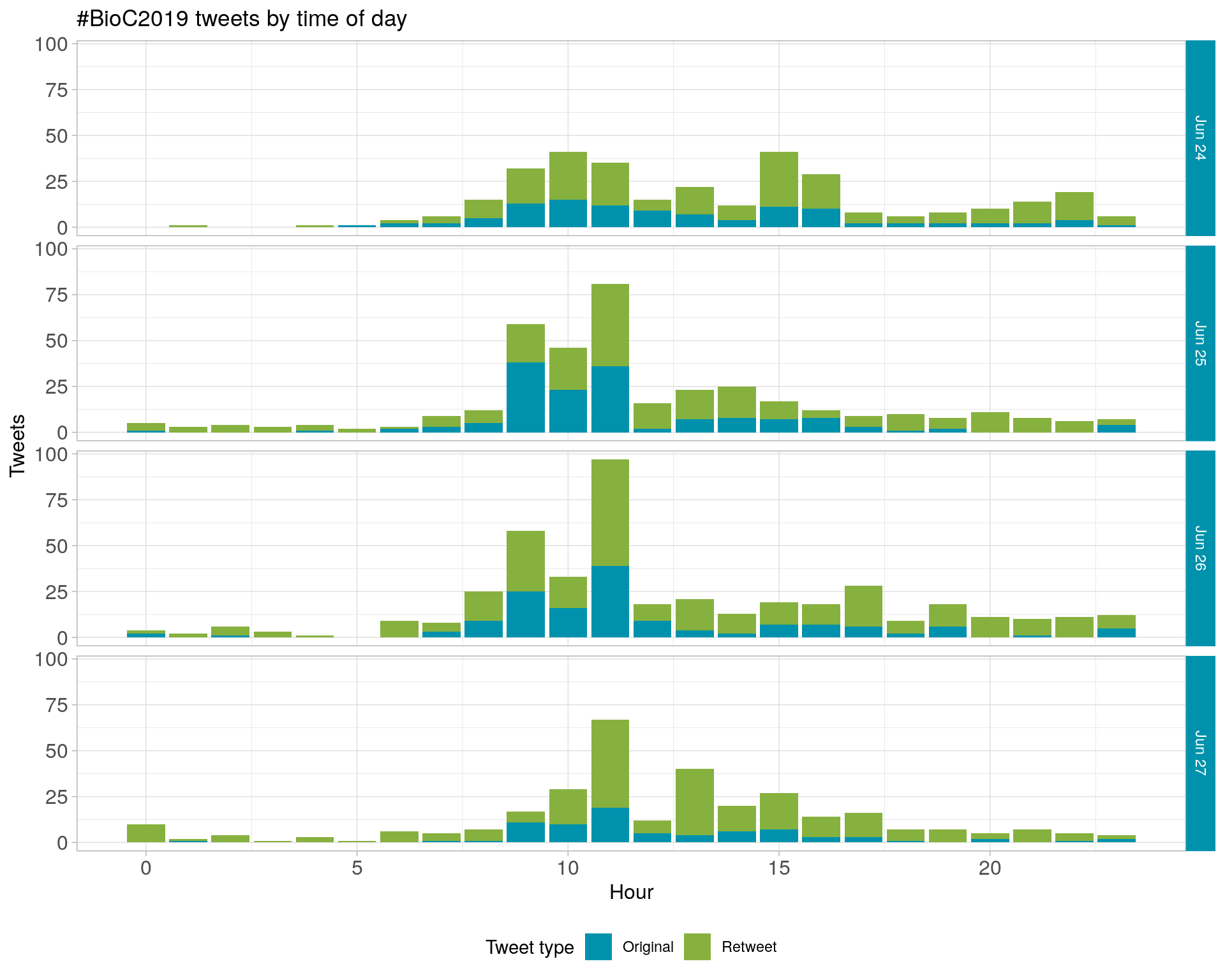
2.2 Tweets by day and time
Filtered for dates 2019-06-24 - 2019-06-27 in the America/New_York timezone.
3 Users
3.1 Top tweeters
Overall
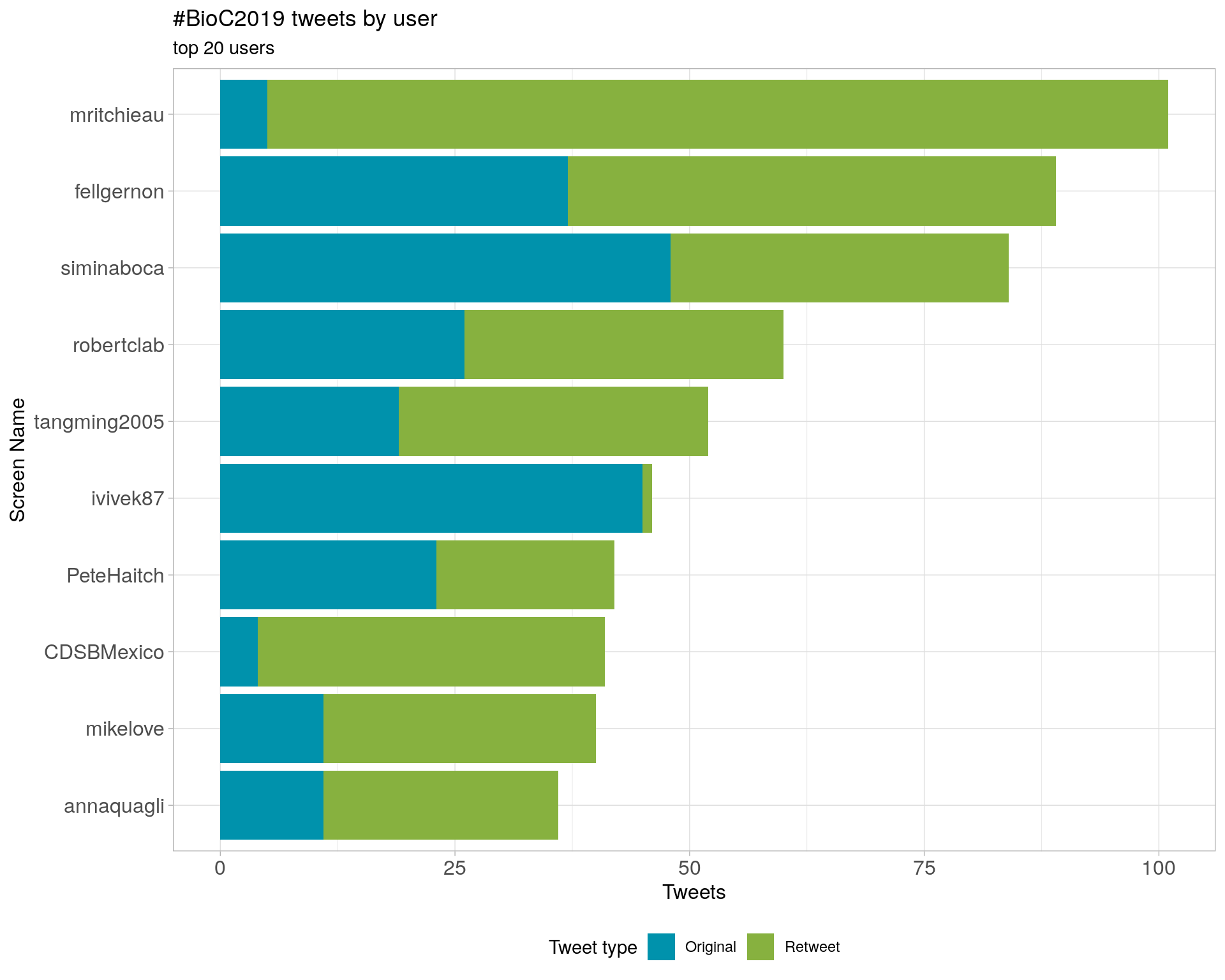
Original
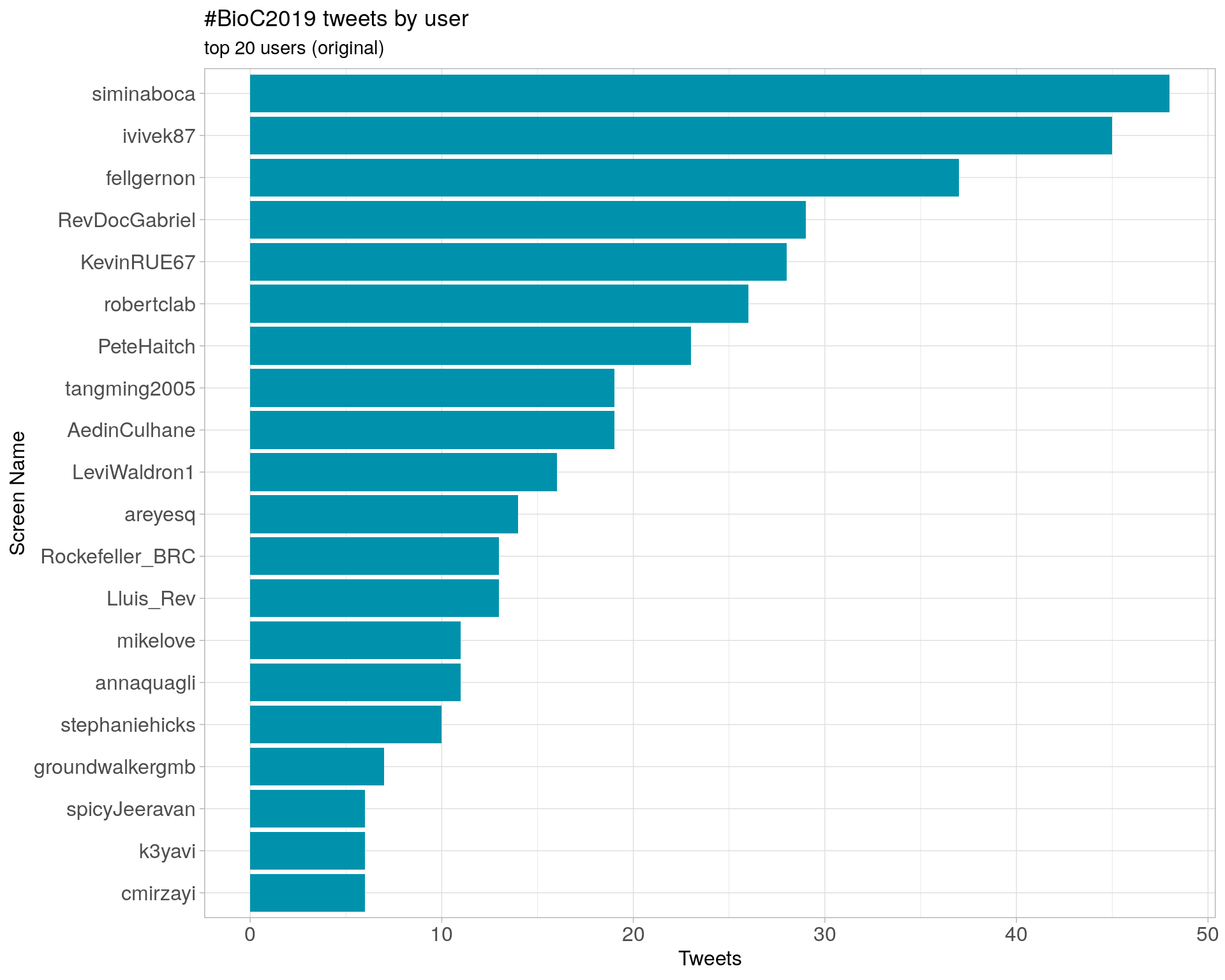
Retweets

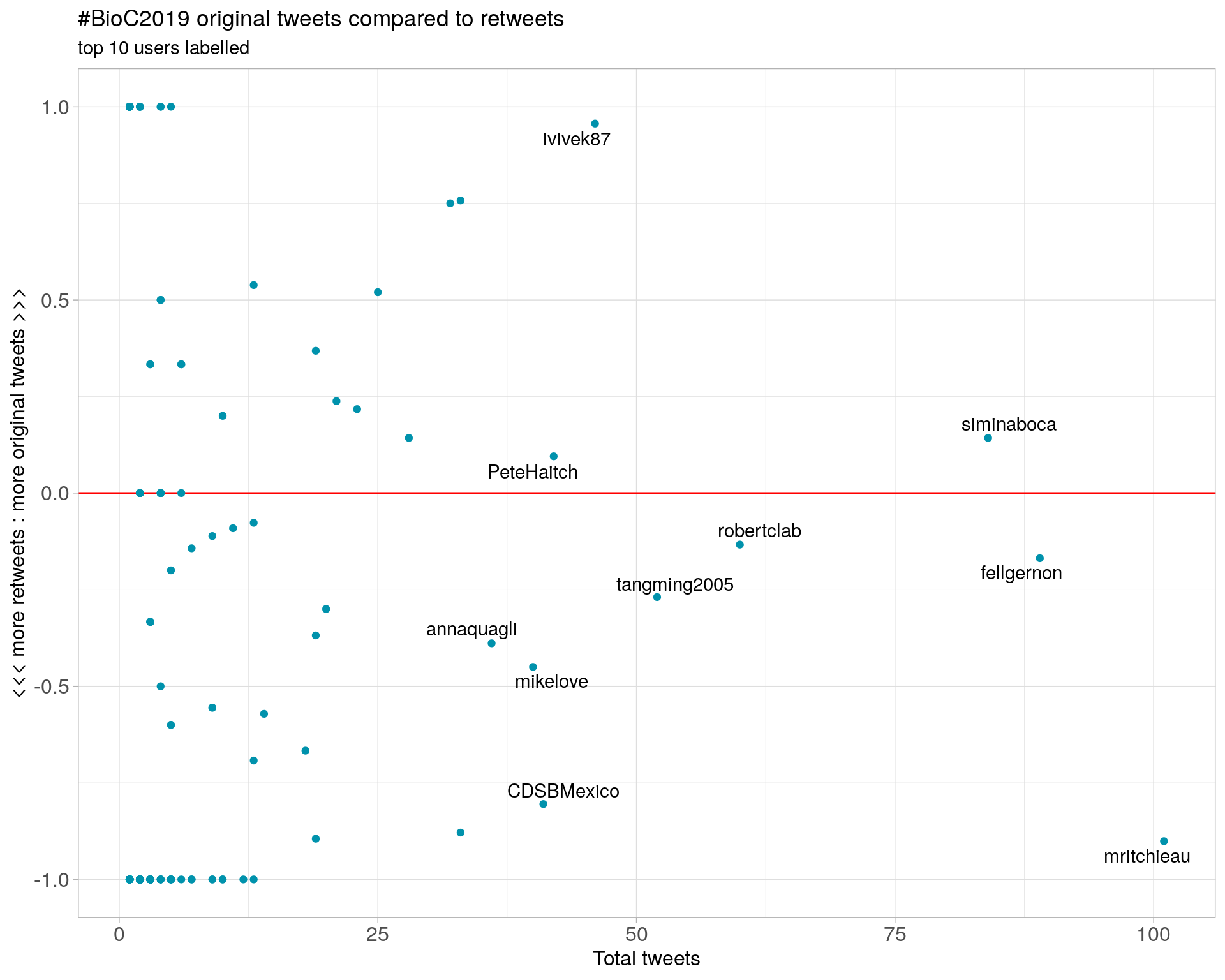
3.2 Retweet proportion
3.3 Top tweeters timeline

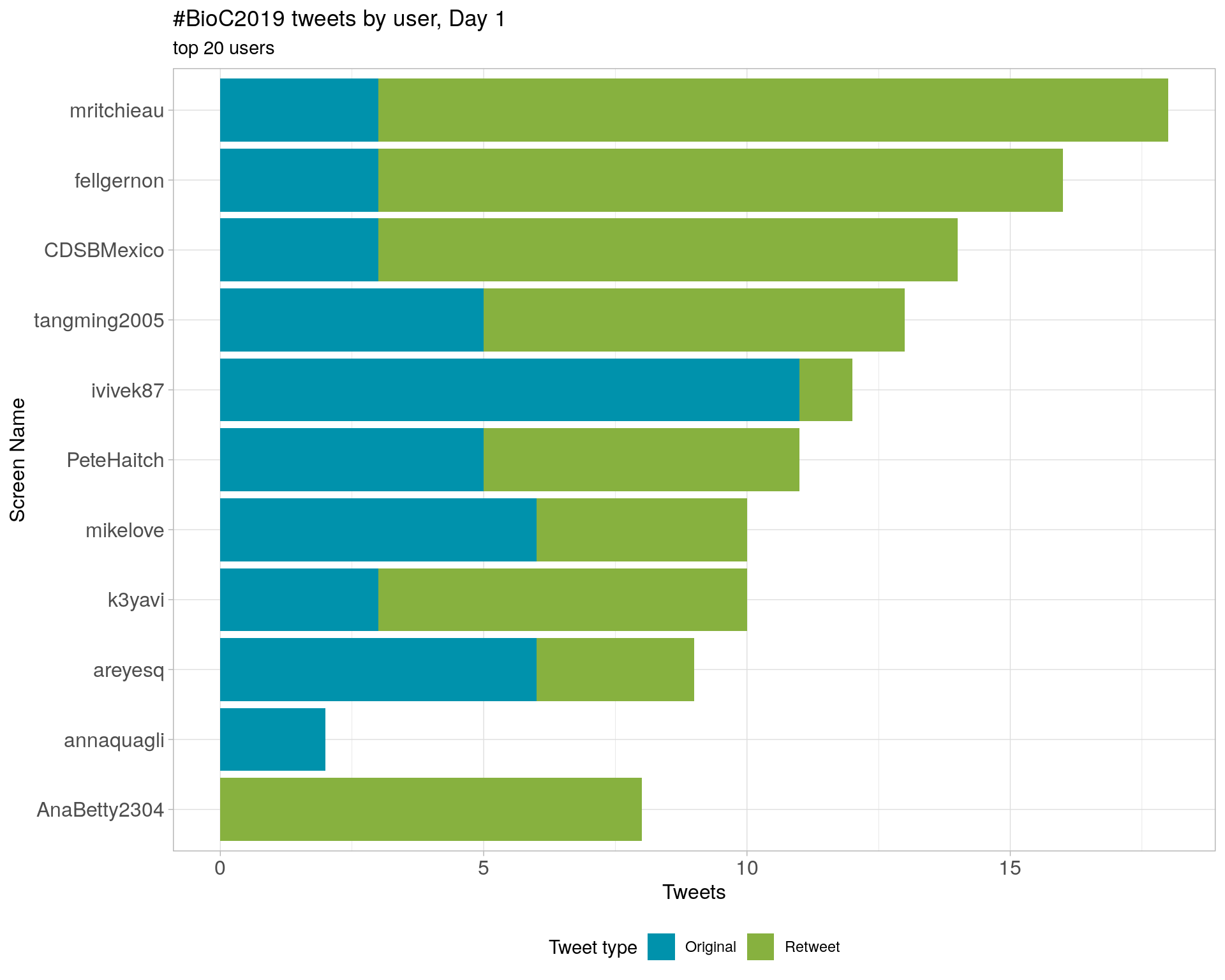
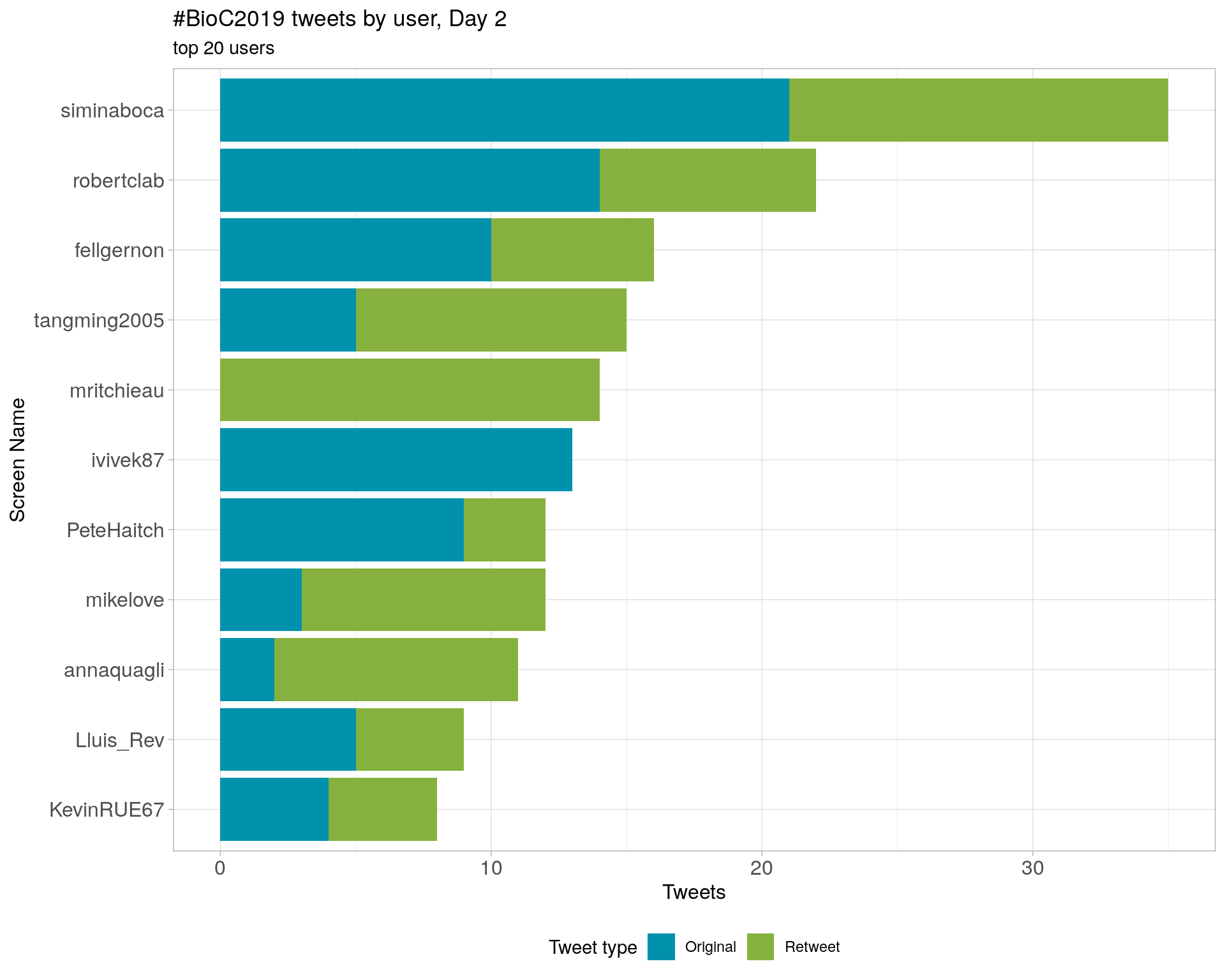
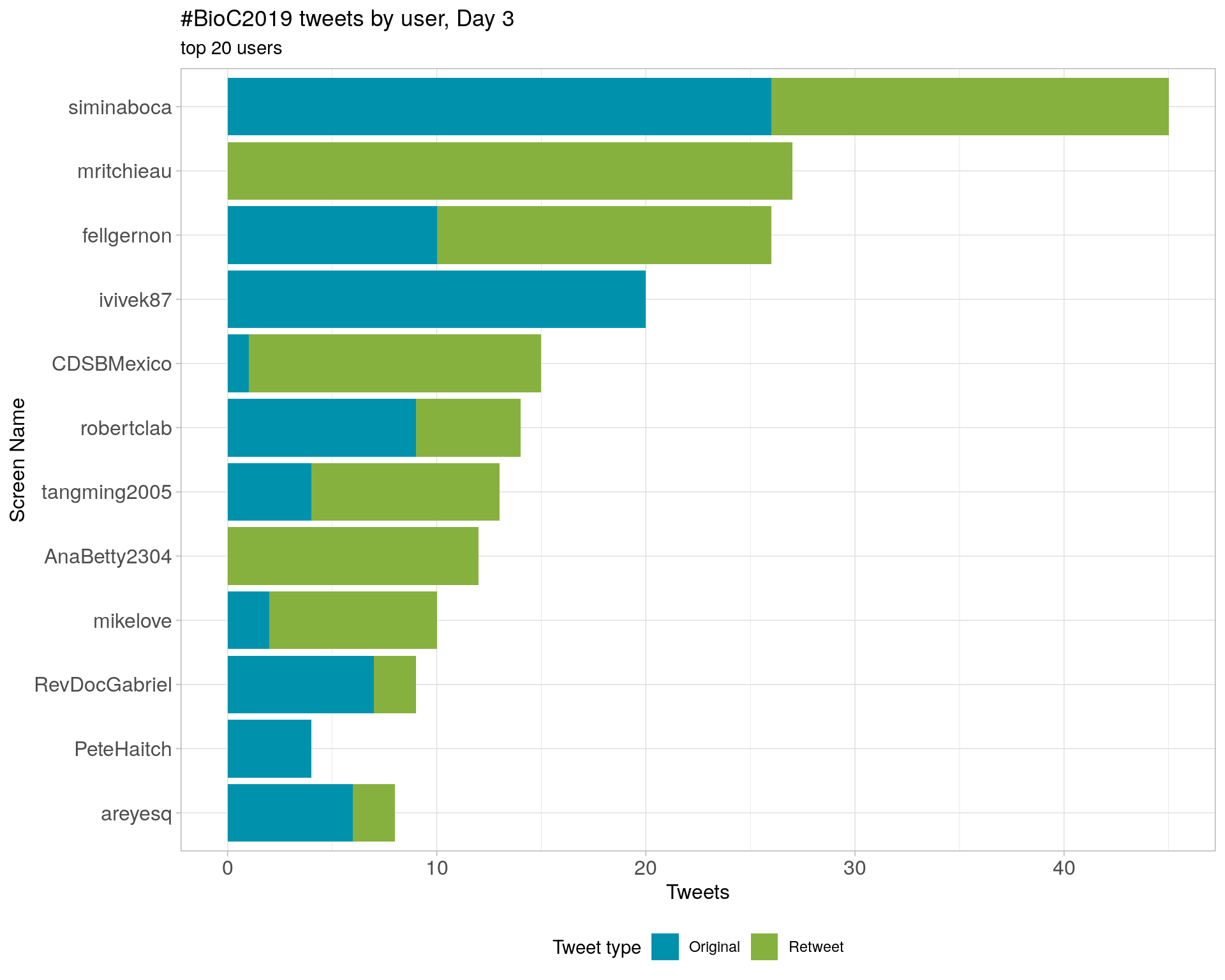
3.4 Top tweeters by day
Overall
Day 1
Day 2
Day 3
Day 4
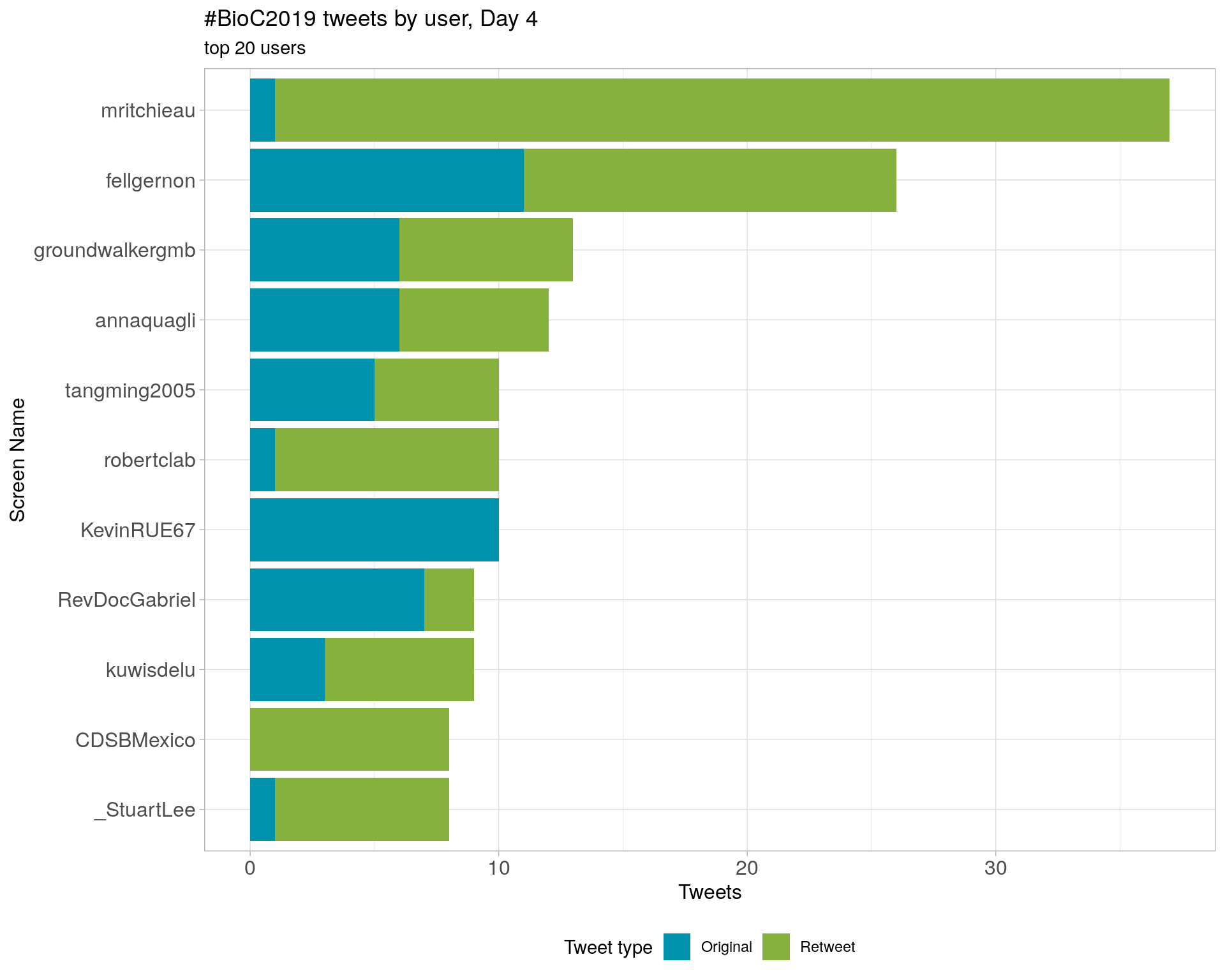
Original
Day 1

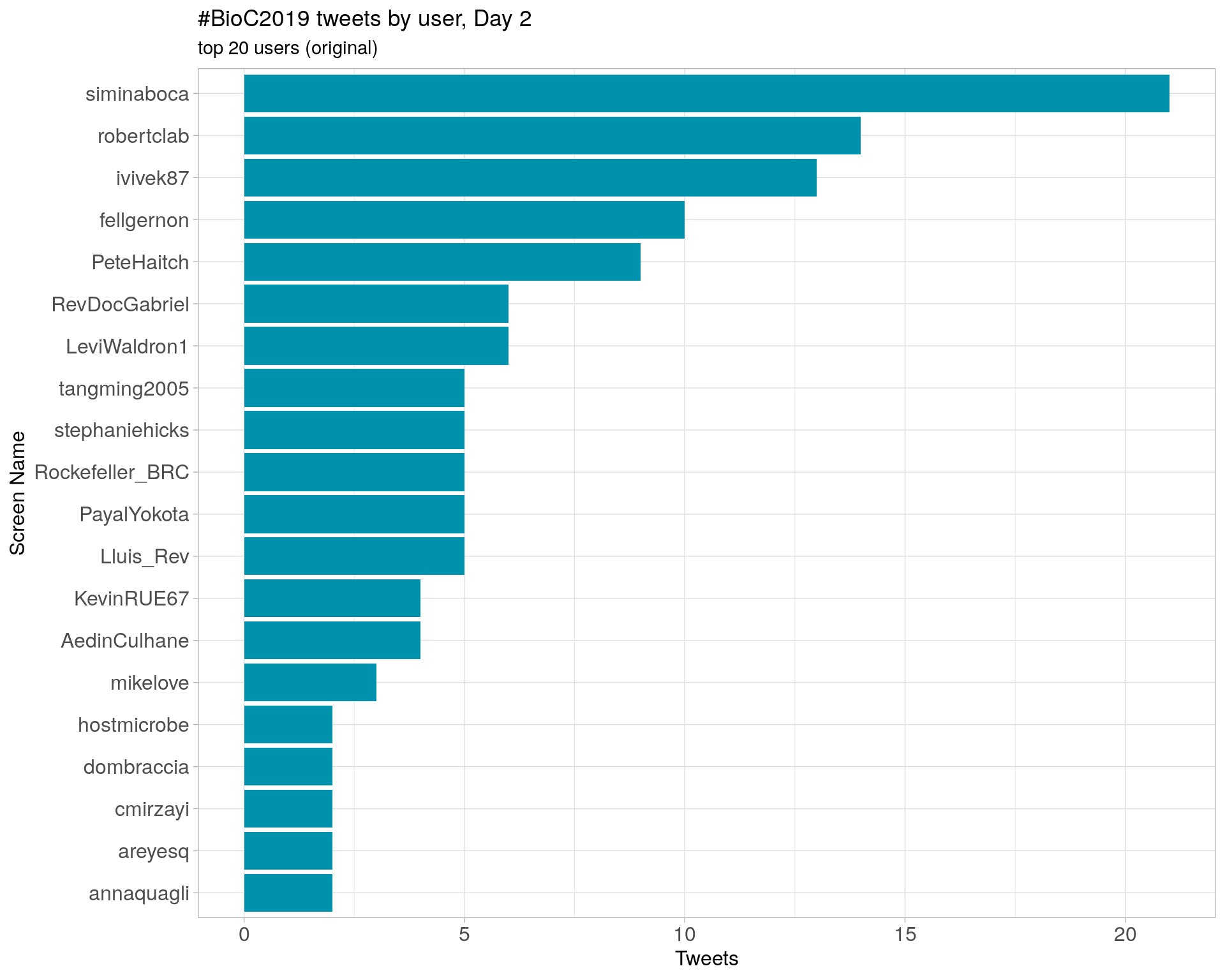
Day 2
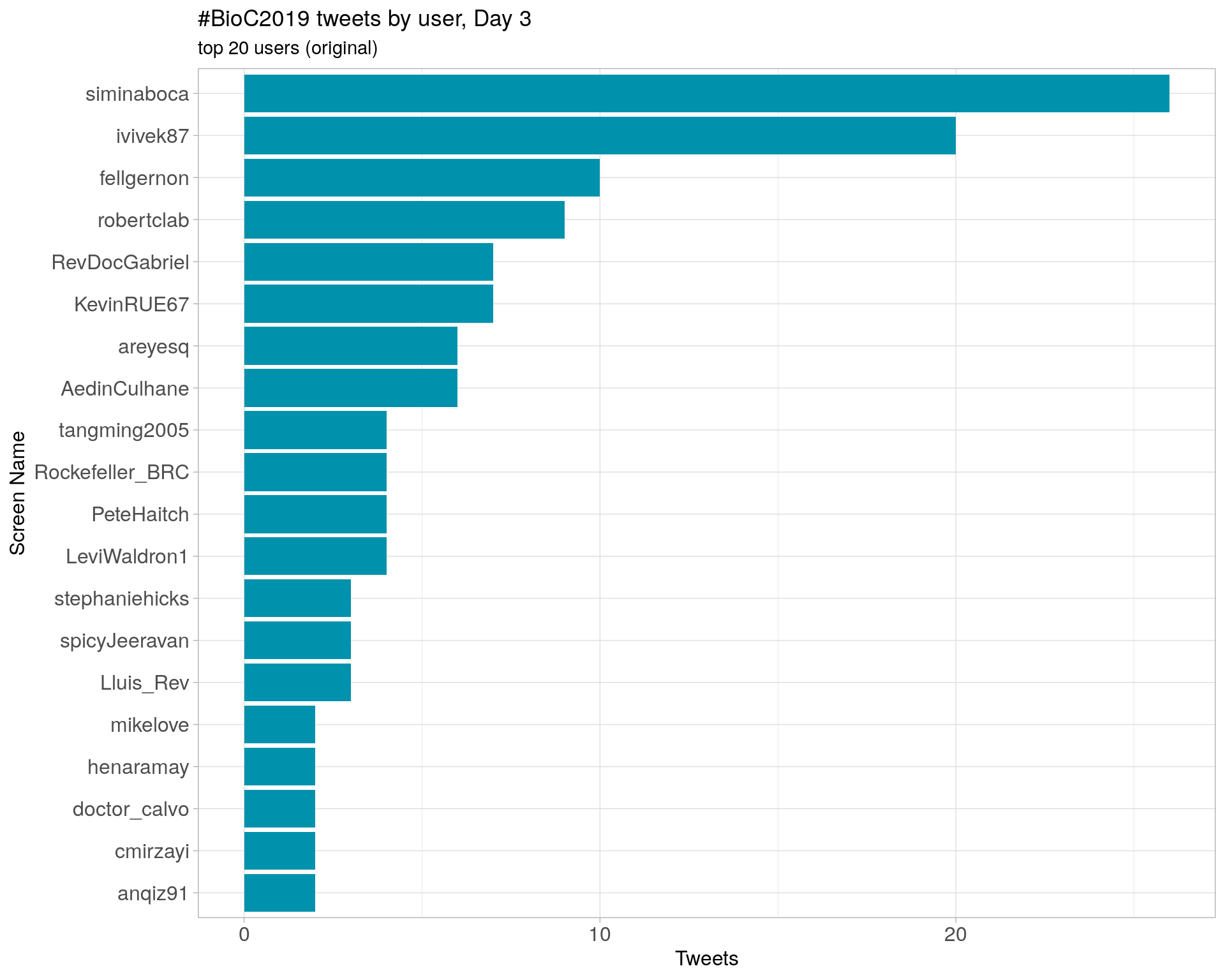
Day 3
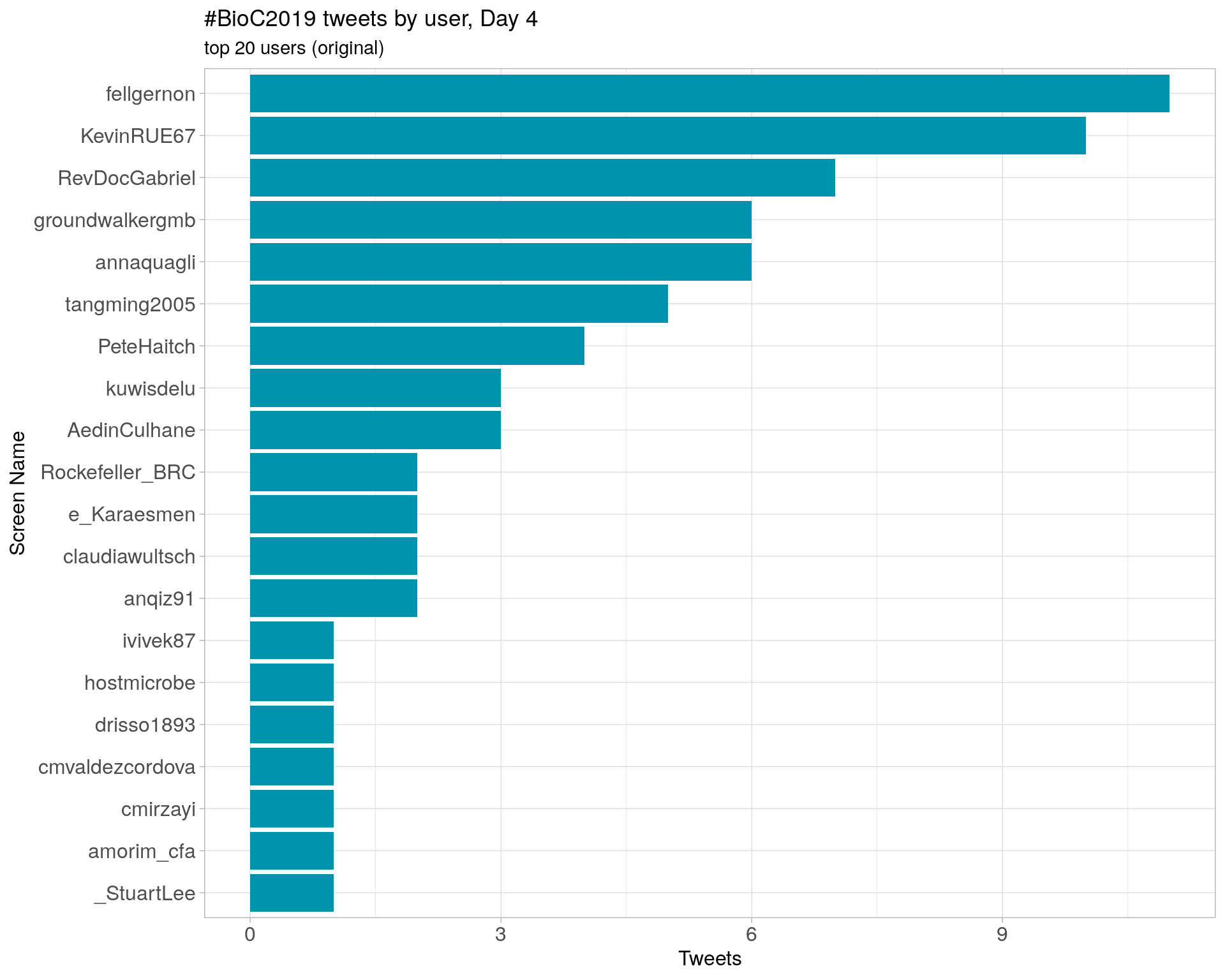
Day 4
Retweets
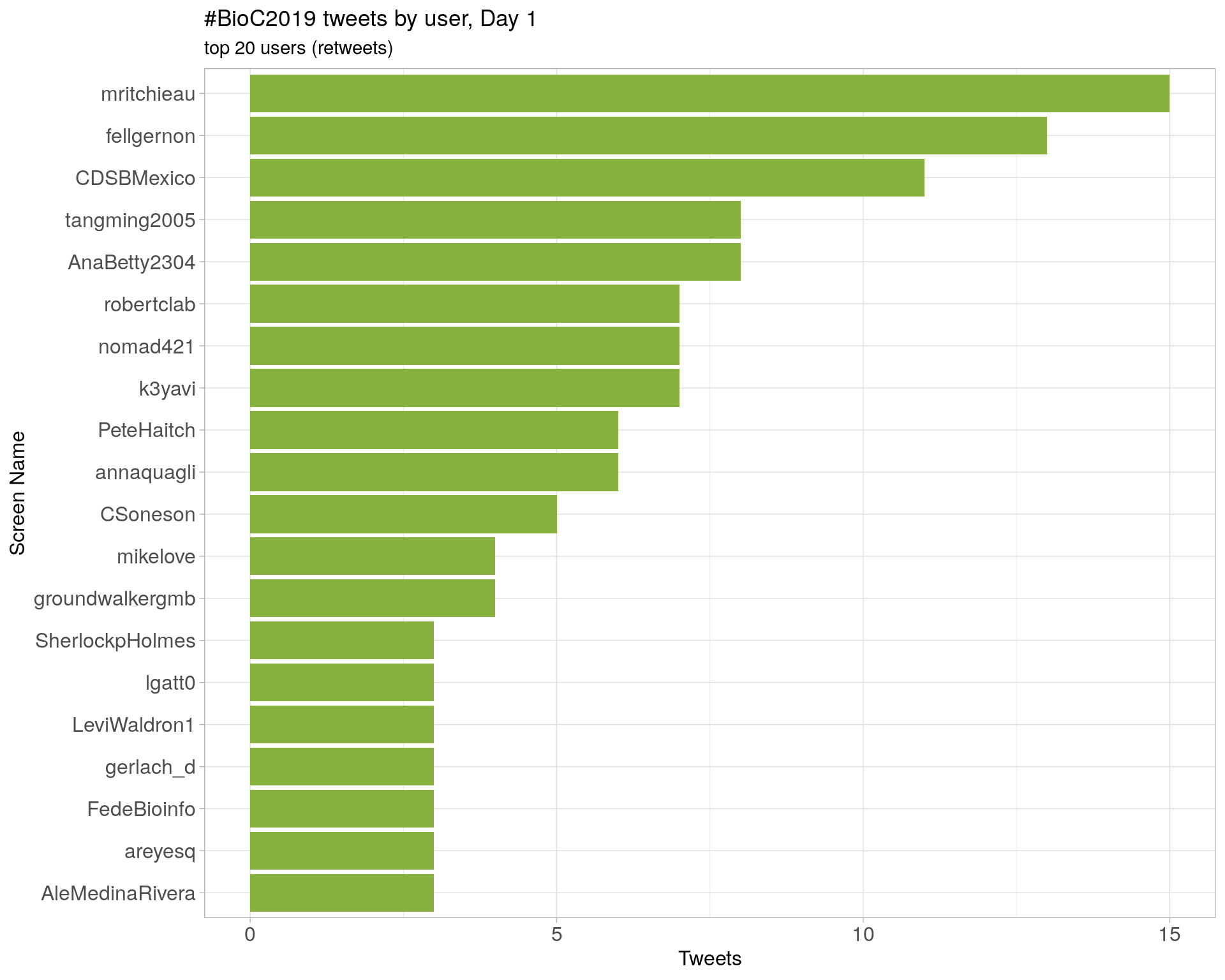
Day 1
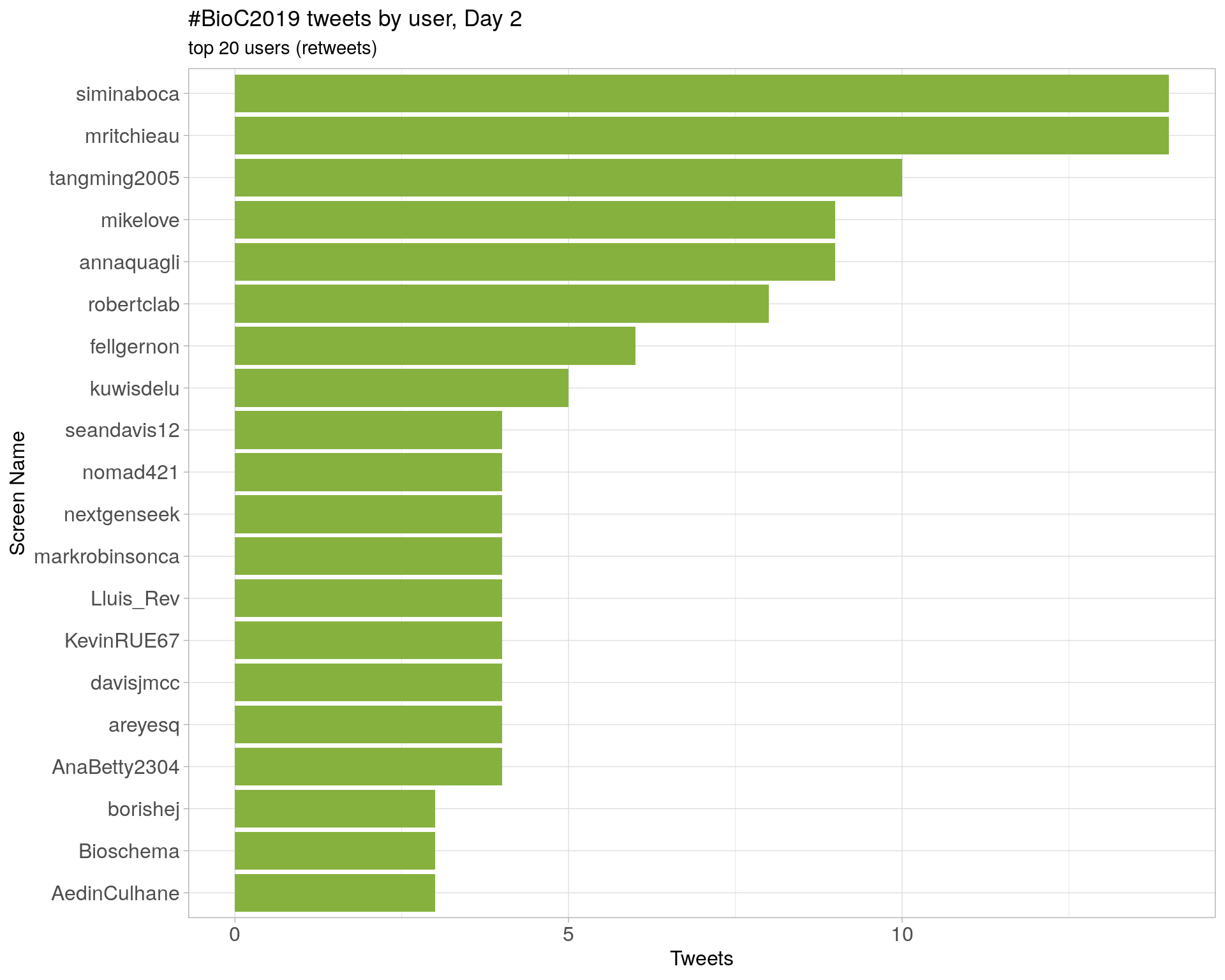
Day 2
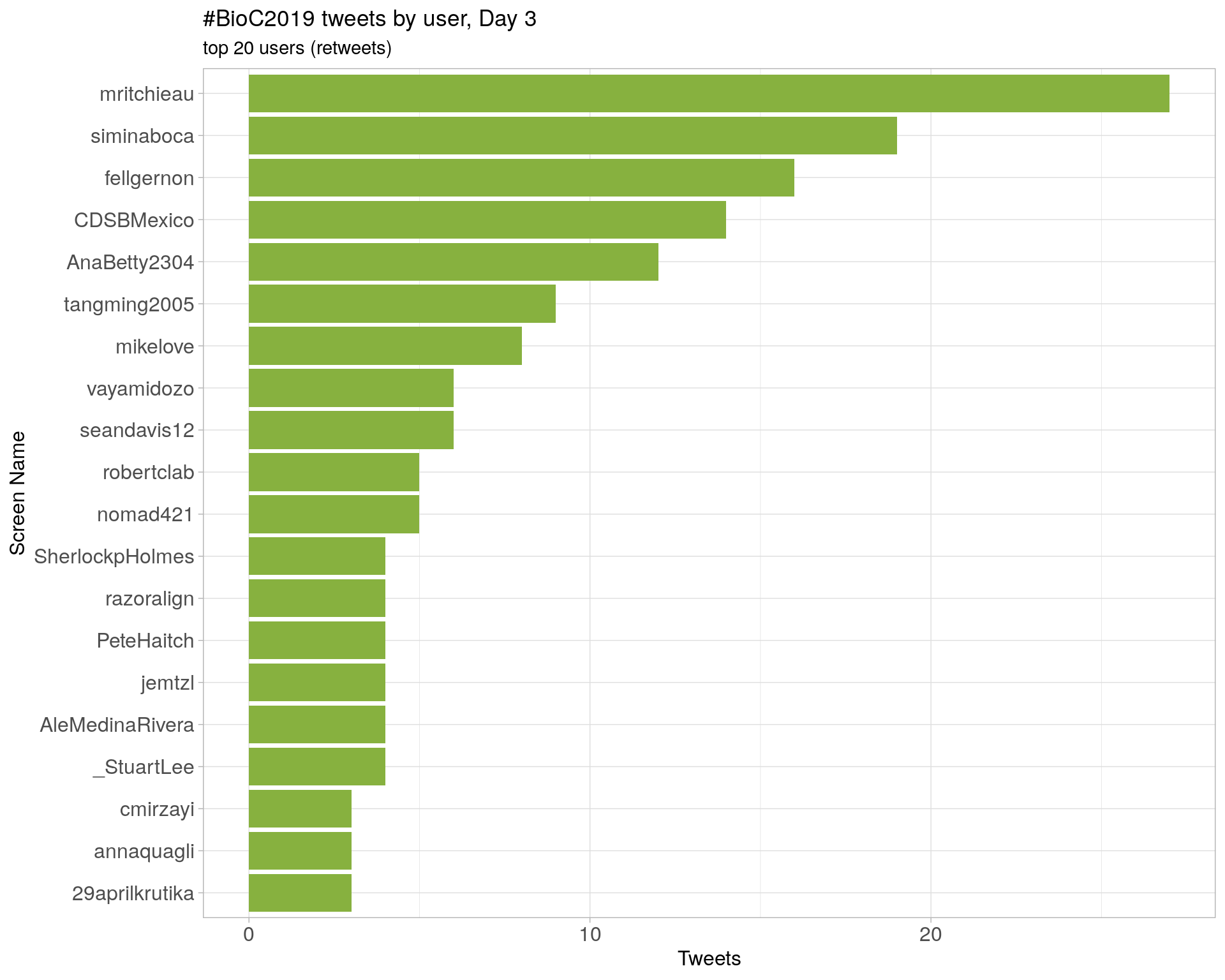
Day 3
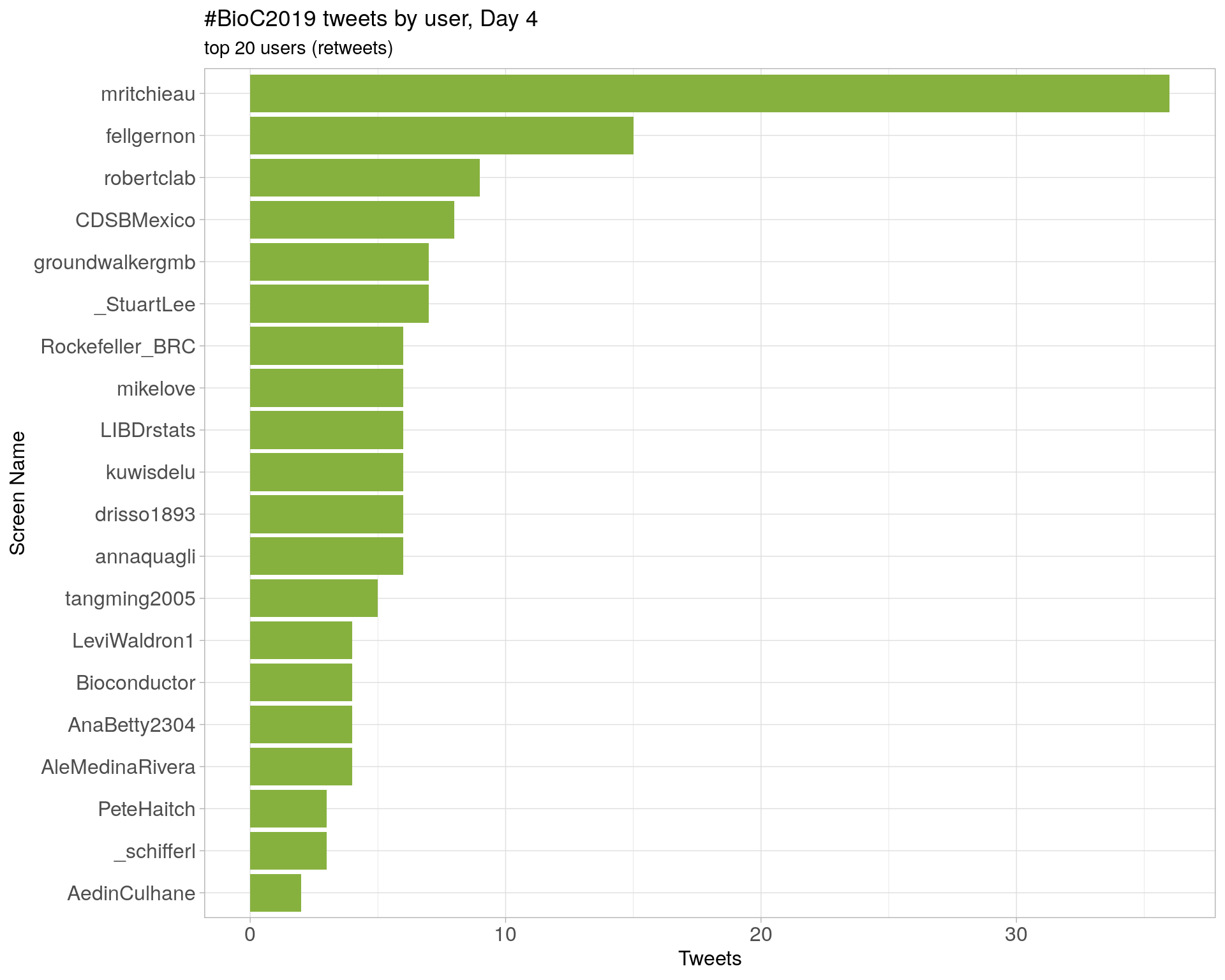
Day 4
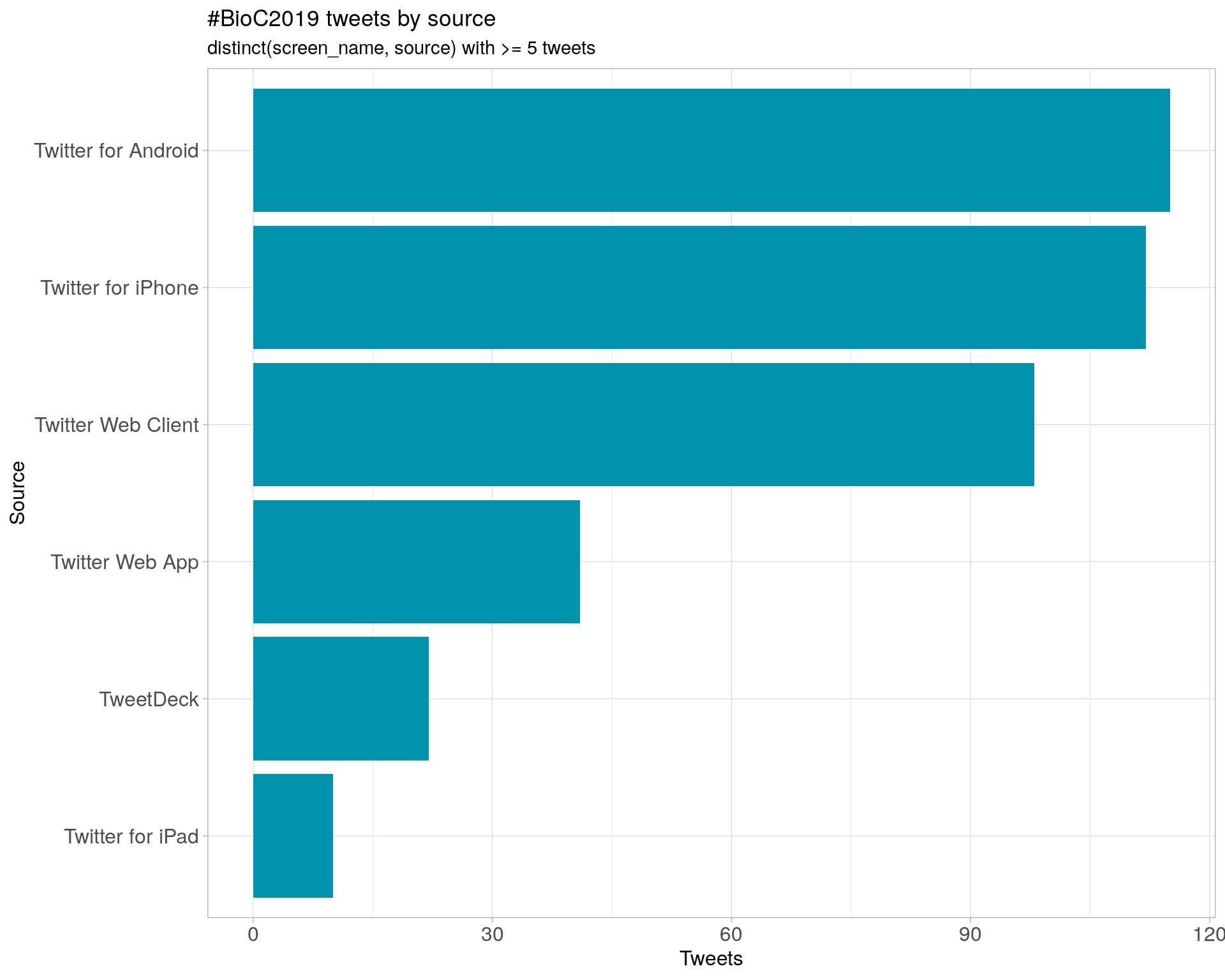
4 Sources
5 Networks
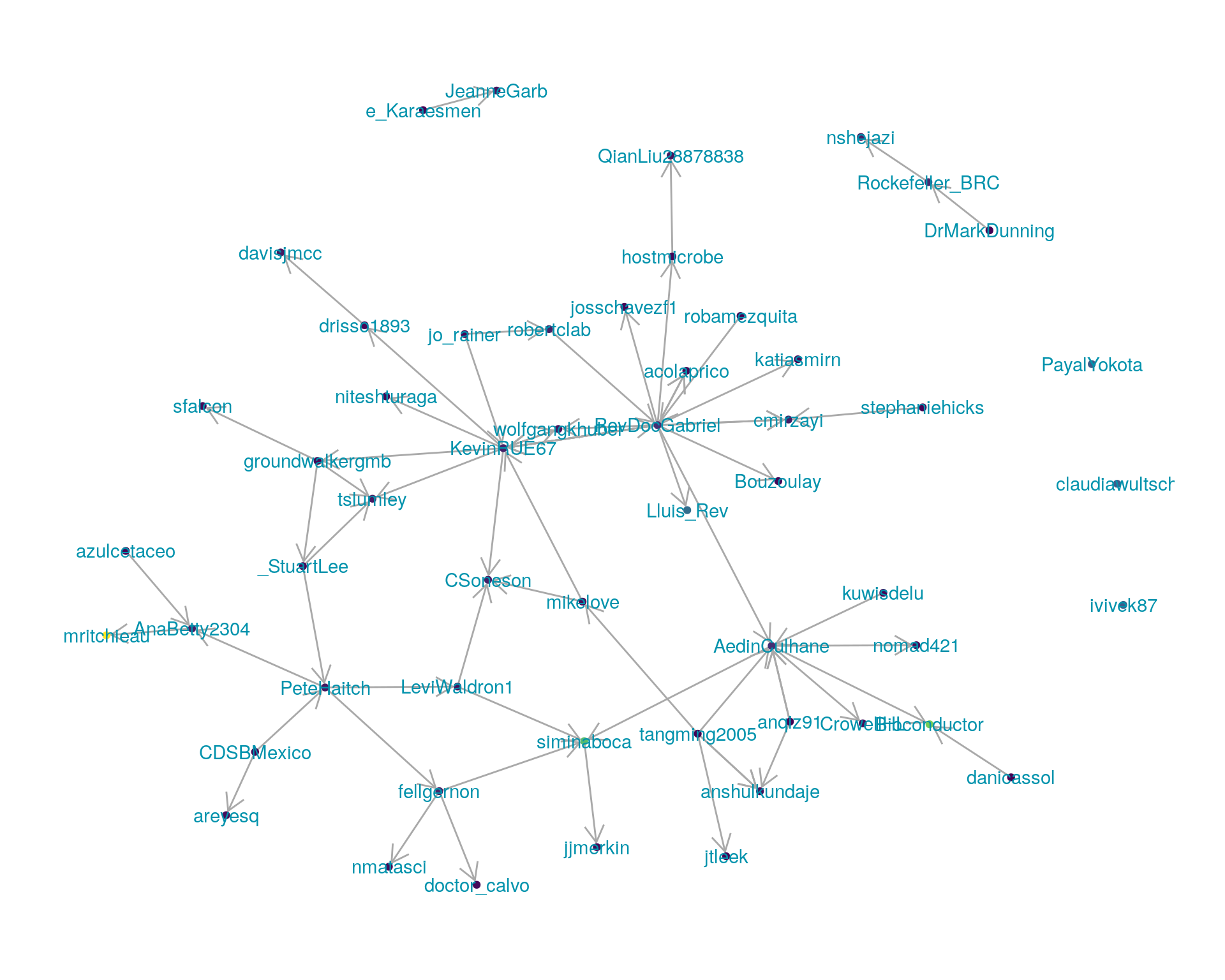
5.1 Replies
The “replies network”, composed from users who reply directly to one another, coloured by PageRank.
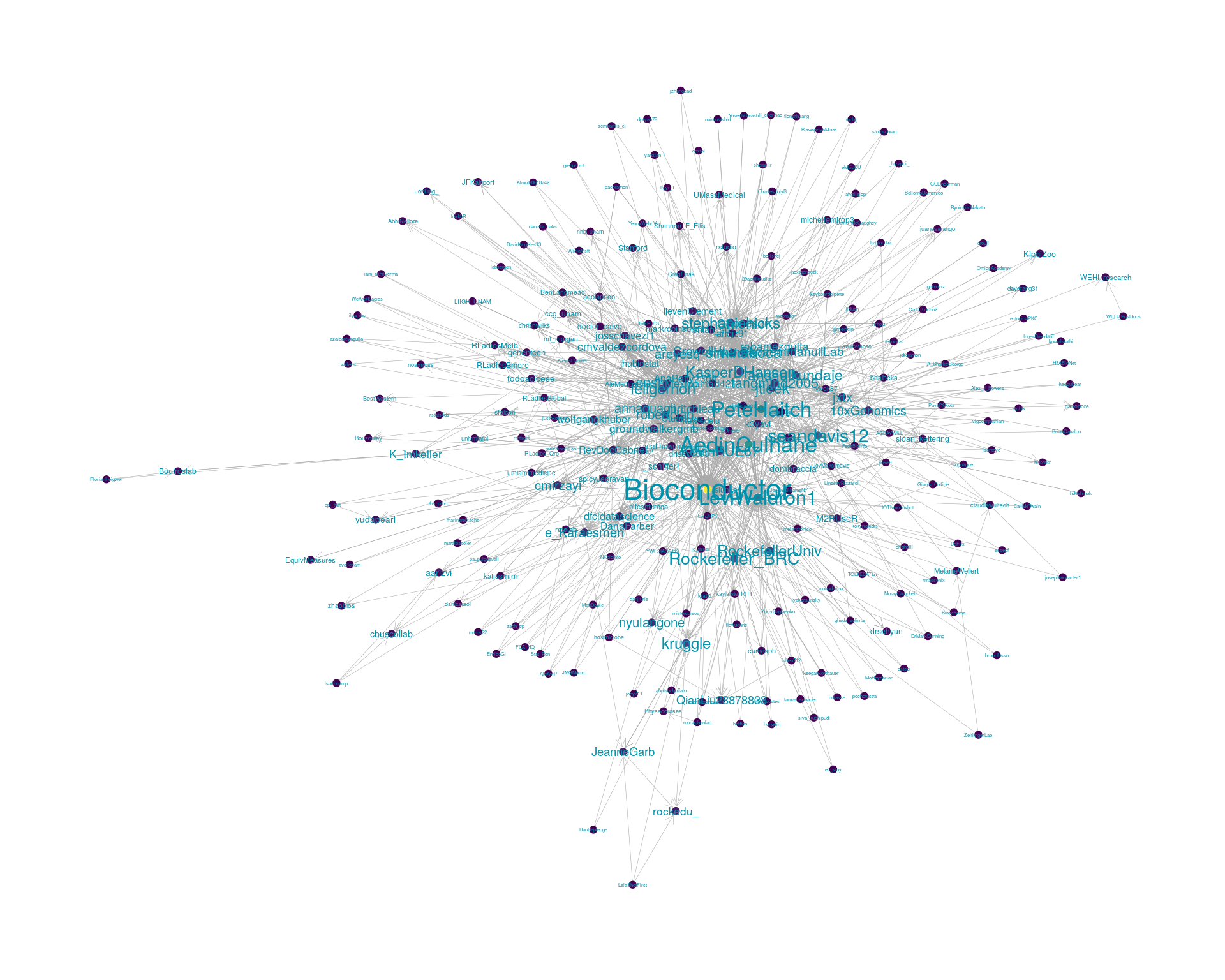
5.2 Mentions
The “mentions network”, where users mention other users in their tweets. Filtered for a k-core of 2. Node colour and size adjusted according to PageRank score.
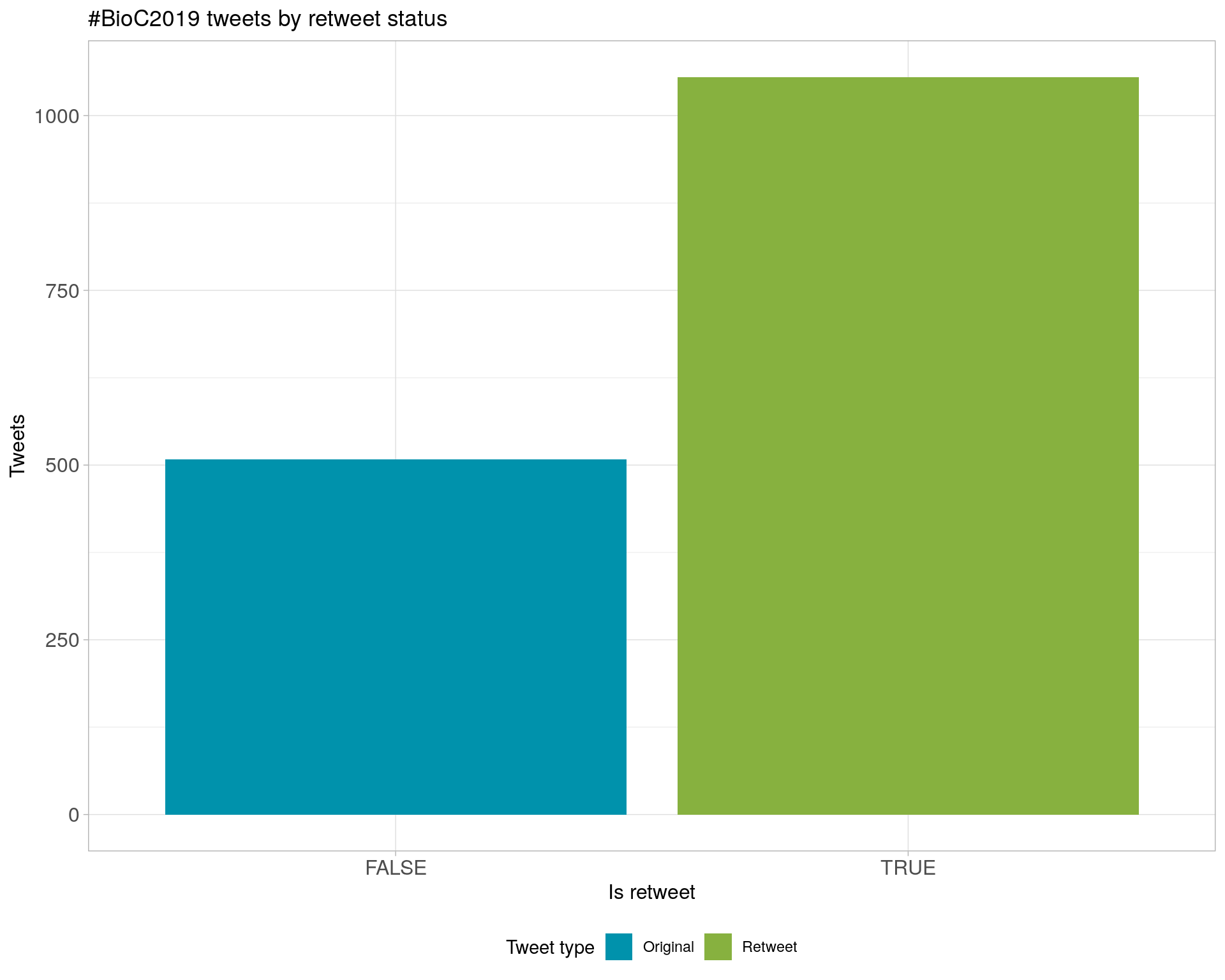
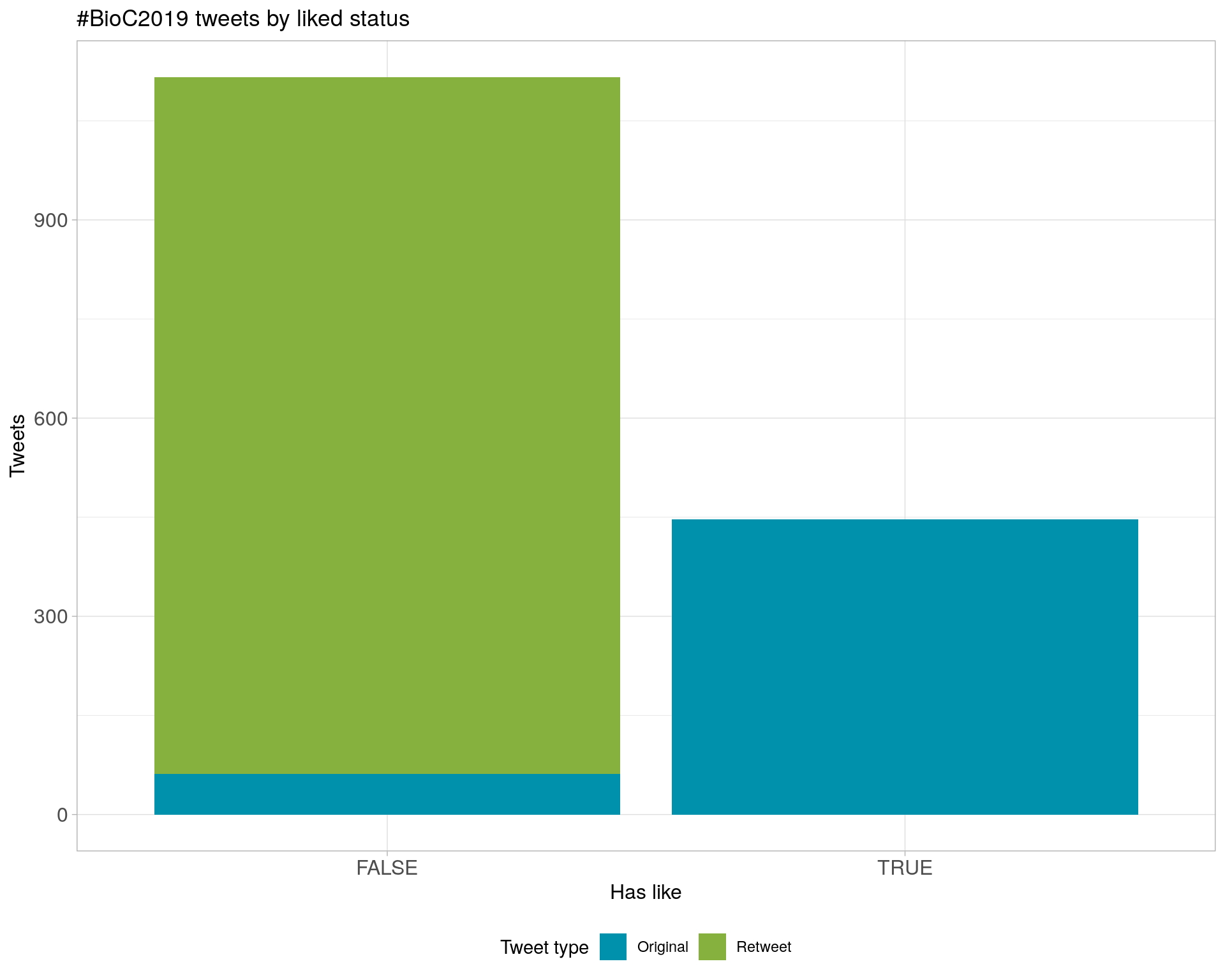
6 Tweet types
6.1 Retweets
Proportion
Count
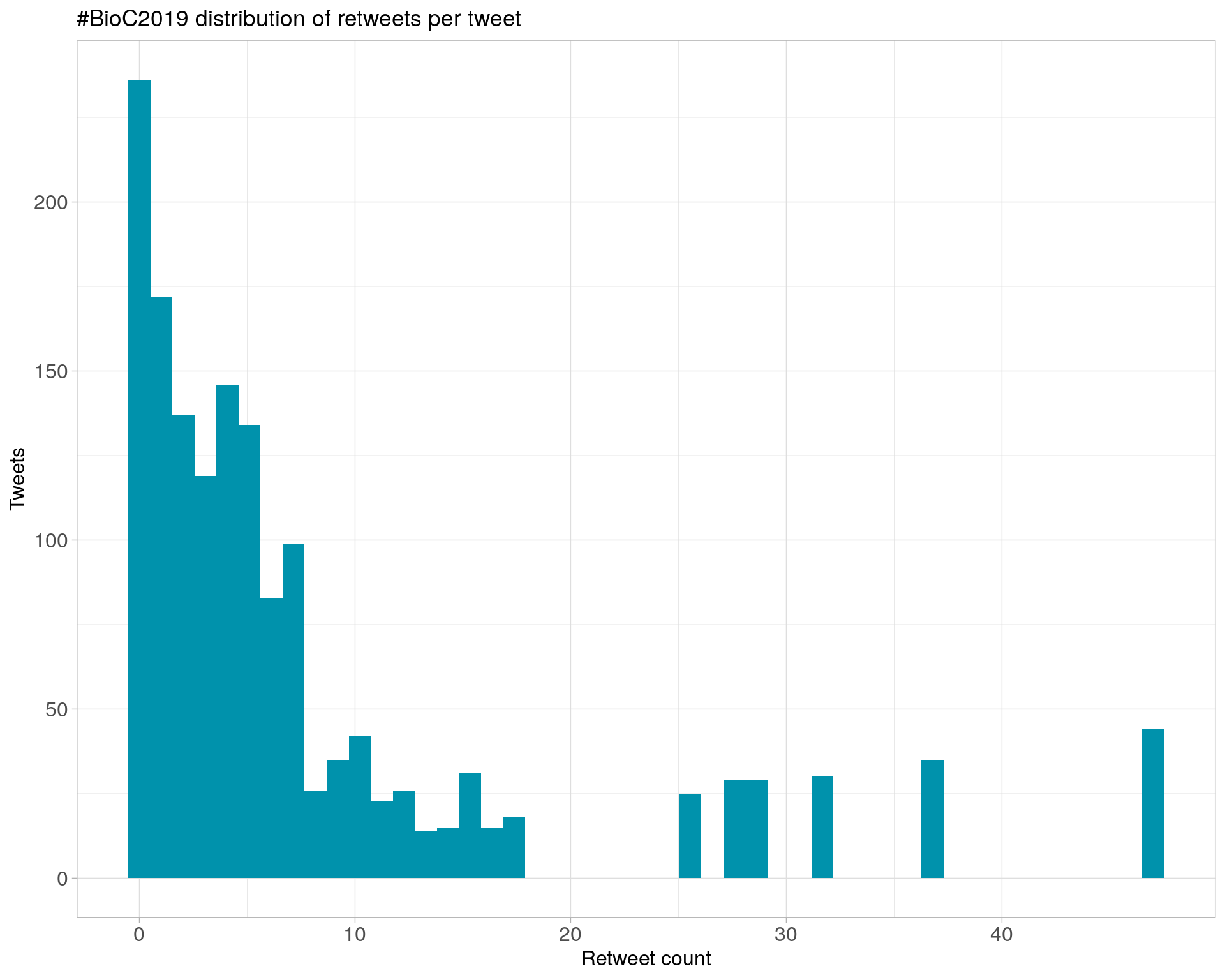
Top 10
| screen_name | text | retweet_count |
|---|---|---|
| seandavis12 | The Bioconductor 2019 conference workshop materials are available online: https://t.co/kpdfKcgjZv. Instructions for use locally are here: https://t.co/59hjl9F36i #BioC2019 https://t.co/xhuuancrl1 | 47 |
| seandavis12 | Just completed (hopefully) final version of the #bioc2019 workshop materials; 26 workshop packages with 607 dependent packages. All available via #docker image (and volume) and Amazon Machine Image. https://t.co/59hjl9F36i Team of folks involved, not to mention workshop authors. | 37 |
| stephaniehicks |
Interested in clustering large #singlecell #rnaseq data with #rstats in a fast and scalable way with data in memory or on-disk (e.g. #hdf5)? Check out my talk in the next section after the break! #BioC2019 📣slides: https://t.co/baaP1dQLIA 💻software: https://t.co/0eBOapo6SJ |
32 |
| LeviWaldron1 | Martin Morgan telling us that @Bioconductor has 1751 packages, 1200 maintainers, and was downloaded by over a half million unique IPs last year. Hello from #bioc2019! https://t.co/pD4D4CuJb5 | 29 |
| KasperDHansen | My lab is hiring post docs (and graduate students). We have multiple collaborators and former lab members at #bioc2019 (unfortunately, I couldn’t attend this year). | 28 |
| PeteHaitch | In case it’s useful, here are my notes from Developer Day at #BioC2019 https://t.co/vkJKiWaT6G (inspired by @jxtx last year) I’ll try to continue over the next few days and write something up following the meeting | 26 |
| PeteHaitch | Robert Gentleman: “If people can’t do things that piss you off then you haven’t given them any power.” (in reference to opening up early #rstats development beyond he and Ross Ihaka and the beginnings of R packages) #BioC2019 | 17 |
| tangming2005 | single cell people, you really should try iSEE https://t.co/BHncVXsNLM awesome package #bioc2019 | 16 |
| robertclab | AK: introducing Kipoi https://t.co/mUW5n46gHi https://t.co/BwQl69U6q5 to facilitate the exchange and reuse of predictive models for genomics #bioc2019 https://t.co/eehcE5gP9l | 15 |
| AedinCulhane | Build an academic website in 10 minutes.. Got no excuse now!! #bioc2019 @bioconductor from @KevinRUE67 #TODOLIST #academiclife https://t.co/I9wcUi8UV1 https://t.co/75zHdN4eCl | 15 |
Most retweeted
6.2 Likes
Proportion
Count

Top 10
| screen_name | text | favorite_count |
|---|---|---|
| seandavis12 | Just completed (hopefully) final version of the #bioc2019 workshop materials; 26 workshop packages with 607 dependent packages. All available via #docker image (and volume) and Amazon Machine Image. https://t.co/59hjl9F36i Team of folks involved, not to mention workshop authors. | 96 |
| LeviWaldron1 | Martin Morgan telling us that @Bioconductor has 1751 packages, 1200 maintainers, and was downloaded by over a half million unique IPs last year. Hello from #bioc2019! https://t.co/pD4D4CuJb5 | 91 |
| tangming2005 | Presenting a poster at #bioc2019 https://t.co/xMxPmENBsX | 87 |
| e_Karaesmen | Awesome to see many #rladies at #BioC2019 maybe we should start #BioCLadies too 😜 https://t.co/vxldiqf6iv | 84 |
| seandavis12 | The Bioconductor 2019 conference workshop materials are available online: https://t.co/kpdfKcgjZv. Instructions for use locally are here: https://t.co/59hjl9F36i #BioC2019 https://t.co/xhuuancrl1 | 67 |
| PeteHaitch | In case it’s useful, here are my notes from Developer Day at #BioC2019 https://t.co/vkJKiWaT6G (inspired by @jxtx last year) I’ll try to continue over the next few days and write something up following the meeting | 64 |
| stephaniehicks |
Interested in clustering large #singlecell #rnaseq data with #rstats in a fast and scalable way with data in memory or on-disk (e.g. #hdf5)? Check out my talk in the next section after the break! #BioC2019 📣slides: https://t.co/baaP1dQLIA 💻software: https://t.co/0eBOapo6SJ |
62 |
| robertclab | Ready for the @Bioconductor meeting #bioc2019 in NYC, bringing along stickers for the two most popular packages I maintain. Reach me out if you want one!! https://t.co/xo5iDqduLo | 61 |
| LeviWaldron1 | In my cancer journal entry today I tried to express how happy I am to be at the #bioc2019 @Bioconductor conference. https://t.co/6ykJw06qFy | 59 |
| mritchieau | On route to #bioc2019 wearing my comfy #user2018 socks. Although my head isn’t quite there yet (jetlag!) my feet are fully prepared :) Presenting 2 workshops on #RNAseq analysis this Wed for those interested https://t.co/htVrPanrqz | 57 |
Most likes

6.3 Quotes
Proportion

Count

Top 10
| screen_name | text | quote_count |
|---|---|---|
| RevDocGabriel | Thanks @LeviWaldron1 for taking the #BioC2019 crew on a tour of #CentralPark #Trailblazer https://t.co/qnB2rtpidU | 3 |
| josschavezf1 | Such a great experience today! I met awesome people at #BioC2019 https://t.co/sMkXCNTBQZ | 3 |
| danicassol | It’s a privilege to be part of this amazing community #bioc2019 #bioconductor https://t.co/OFIKL5XzAr | 3 |
| amorim_cfa | As a biologist, the world of bioinformatics is attracting more and more - that’s what I want to do. Just came back from #BioC2019, excellent experience, talks, networking… it was an amazing conference! #Rladies #NewYork #bioinformatics https://t.co/Z7wX5t2l9t | 3 |
| e_Karaesmen | @JeanneGarb Oh I think you missed us because we were taking a photo… As a female PhD student I think @Bioconductor is one the most inclusive tech communities in the world. #BioC2019 https://t.co/ft1u7XT4a0 | 3 |
| annaquagli | And great to have spread the word to new ones who had never heard about #rladies @RLadiesGlobal #BioC2019 https://t.co/cHbjuvkxJW | 3 |
| ivivek87 | This is one conference I wish to attend in future. #BioC2019 seem 🔥. Twitter is all I have to learn about it this year. Bring it on. Something I learnt early on. Don’t reinvent wheel if you Jane options, one can work their way through if one digs deep. # @Bioconductor speaks! https://t.co/sabVIm0cyC | 2 |
| claudiawultsch | Excited to be joining in tomorrow #BioC2019 #Bioconductor https://t.co/YCQZa2aRfs | 2 |
| PeteHaitch | Argh, after the break we have parallel sessions! Anyone attending the ‘Statistical methods’ section want to take notes? #Bioc2019 https://t.co/9XKcFo6DZU | 2 |
| UofABioinfoHub | Here’s the URL from Pete’s notes in case it gets hidden on a retweet: https://t.co/RAqXSuKFVj #BioC2019 https://t.co/YQibd6RnpV | 2 |
Most quoted

7 Media
Proportion

Top 10
| screen_name | text | favorite_count |
|---|---|---|
| LeviWaldron1 | Martin Morgan telling us that @Bioconductor has 1751 packages, 1200 maintainers, and was downloaded by over a half million unique IPs last year. Hello from #bioc2019! https://t.co/pD4D4CuJb5 | 91 |
| tangming2005 | Presenting a poster at #bioc2019 https://t.co/xMxPmENBsX | 87 |
| e_Karaesmen | Awesome to see many #rladies at #BioC2019 maybe we should start #BioCLadies too 😜 https://t.co/vxldiqf6iv | 84 |
| seandavis12 | The Bioconductor 2019 conference workshop materials are available online: https://t.co/kpdfKcgjZv. Instructions for use locally are here: https://t.co/59hjl9F36i #BioC2019 https://t.co/xhuuancrl1 | 67 |
| robertclab | Ready for the @Bioconductor meeting #bioc2019 in NYC, bringing along stickers for the two most popular packages I maintain. Reach me out if you want one!! https://t.co/xo5iDqduLo | 61 |
| mritchieau | On route to #bioc2019 wearing my comfy #user2018 socks. Although my head isn’t quite there yet (jetlag!) my feet are fully prepared :) Presenting 2 workshops on #RNAseq analysis this Wed for those interested https://t.co/htVrPanrqz | 57 |
| QianLiu28878838 | Bioconductor team is celebrating the success of 15th conference in NYC! #BioC2019 https://t.co/2MlfXxkZji | 51 |
| AedinCulhane | @Bioconductor #bioc2019 in #NYC https://t.co/m5qbaQpXd6 | 42 |
| michellemiron3 | Avi presents a tool he created for estimating gene abundance from single-cell RNA seq data #bioc2019 https://t.co/nXBr2dNwCW | 41 |
| LeviWaldron1 | New Waldron Lab photo at #bioc2019! I couldn’t be prouder of a group that works so happily and productively together, or of the four besides me who presented in workshops this year. We contributed: (1/2) @cmirzayi @drsehyun @M2RUseR & alumnus @_schifferl. https://t.co/6EwYcadr2L https://t.co/oCFZipEPCy | 40 |
7.1 Most liked image

8 Tweet text
8.1 Word cloud
The top 100 words used 3 or more times.

8.2 Bigram graph
Words that were tweeted next to each other at least 3 times.

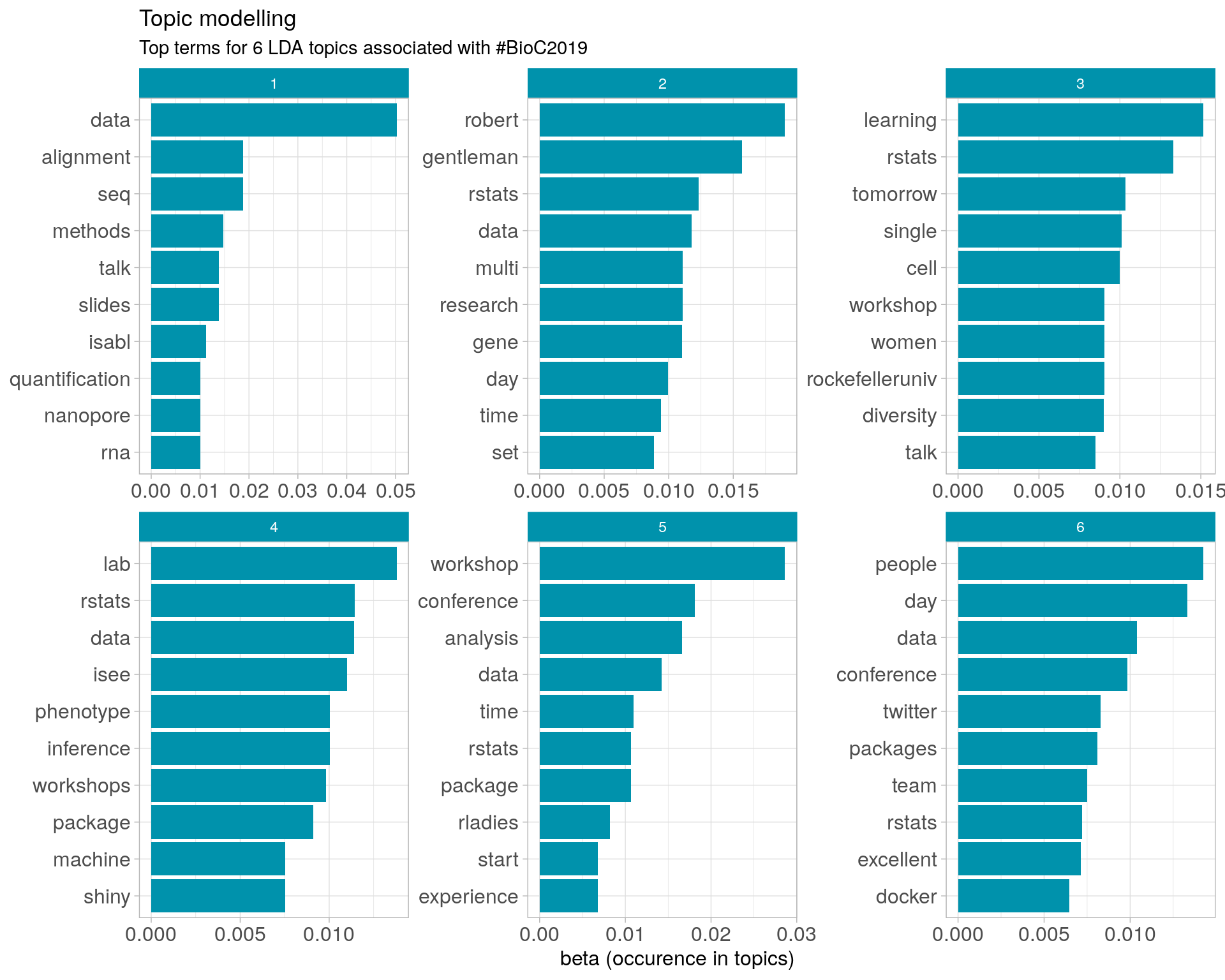
8.3 Topic modelling
Top 10 words associated with 6 topics identified by LDA.
8.3.1 Representative tweets
Most representative tweets for each topic
Topic 1
| screen_name | text | gamma |
|---|---|---|
| PapaemmanuilLab | Real pleasure to introduce #Isabl at #bioc2019 this am. Very gratifying to see the response! Structured data architectures as digital biobanks, secure data governance, reproducibility and foster data sharing, analyses and innovation. Working towards Isabl release soon #staytuned https://t.co/sPMGfToWTk | 0.9946292 |
| siminaboca | @anshulkundaje AK is modeling transcription factor data from ChIP-seq data or chromatin accessibility data from DNAase-seq or ATAC-seq. How do we predict if individual sites are active or inactive in terms of transcription binding? #BioC2019 | 0.9943973 |
| LeviWaldron1 | Nathan Sheffield presents a well-engineered solution to data provenance in the metadata slot of @Bioconductor objects. You use a single yaml file containing locations of your source data file + annotations, and R script to load data https://t.co/IQzBPUwhiY #BioC2019 https://t.co/Bh8PSyq8hf | 0.9941444 |
| AedinCulhane | SA. New seq alignment which is halfway between salmon quasi and full alignment approach. Towards selective-alignment: Bridging the accuracy gap between alignment-based and alignment-free transcript quantification | bioRxiv #bioc2019 https://t.co/wwfoCZoOO8 | 0.9938676 |
| stephaniehicks |
Interested in clustering large #singlecell #rnaseq data with #rstats in a fast and scalable way with data in memory or on-disk (e.g. #hdf5)? Check out my talk in the next section after the break! #BioC2019 📣slides: https://t.co/baaP1dQLIA 💻software: https://t.co/0eBOapo6SJ |
0.9935634 |
| k3yavi | #bioc2019 the effect on quantification estimates based on the different alignment/mapping methods used on “real” data, compared to hybrid Oracle . The problem of RNA-seq quantification is not solved… https://t.co/qtsVMhj9aa | 0.9924378 |
| LeviWaldron1 | Key slides from Nitesh Turaga’s presentation on Docker for easy dev, use, and for creating clusters on the cloud called from R session. Slides at https://t.co/Go7KW4uChu #BioC2019 #Docker https://t.co/pwlQmhzFBQ | 0.9924378 |
| siminaboca | @PapaemmanuilLab EP describes ISABL, a platform for scalable biostat informatics analysis - includes RESTful API, integrated with institutional RedCap, can run everything on laptop in 10 minutes #bioc2019 | 0.9924378 |
| robertclab | CS: applied different quantification methods to nano-ore and illumina data and both technologies provide comparable correlations between replicates. So, yes, nanopore data is quantitative #bioc2019 | 0.9919697 |
| robertclab | At the Robert Gentleman’s 60th Birthday Symposium, Richard Bourgon from @genentech gives a definition of what is a computational biologist: “computational is a modifier, we’re biological scientists” #bioc2019 https://t.co/zSINCGFWI5 | 0.9919697 |
Topic 2
| screen_name | text | gamma |
|---|---|---|
| fellgernon |
Martin Morgan @mt_morgan (@Bioconductor lead) at the Robert Gentleman symposium “We are still using S4” /jk Last yr 500k unique IP for downloads 30k PubMed citations (vs ~1.4k when MM took over) RG gave him the keys to THE car, not a Ferrari, but still the car! 😱 #BioC2019 |
0.9946292 |
| siminaboca | @AedinCulhane AC: moGSA is a PCA-based method and can also be use for single sample GSEA (ssGSEA). She applied it to finding PanCancer immune subtypes in TCGA data by generating ssGSEA scores for immune sets and pathways and then clustering them. #BioC2019 | 0.9943973 |
| fellgernon | Pete @PeteHaitch is teaching us about the #DelayedArray framework developed by Hervé Pàges from @Bioconductor and now is showing us his #DelayedMatrixStats 📦 which enables matrix statistical functions on HDF5 data matrices. Motivated by #scRNAseq and #DNAm #WGBS data #BioC2019 https://t.co/FG8jTtNQvI | 0.9938676 |
| mritchieau | To save time, I’ve reformatted my lightning talk as a tweet! If you’re a package developer, consider creating a #hexsticker and add it to the growing collection at https://t.co/mrwPKBnhMR. For some cool examples see 👇 #bioc2019 https://t.co/VMXc4xeOud | 0.9924378 |
| RevDocGabriel | Such an honour for two avid learnRs from @umiamimedicine to meet the grandmaster of R himself, Prof. Robert Gentleman, at #BioC2019. Thank you, @Bioconductor, for this wonderful opportunity and the awards to travel to @RockefellerUniv. We will be back next year! #Rstats #R4DS https://t.co/7R9axo7dBS | 0.9924378 |
| anshulkundaje | #bioc2019 community - we’d love to have u involved in contributing to, developing, using, providing feedback about @KipoiZoo - ML model zoo for genomics. We’re inspired by the amazing @Bioconductor community. https://t.co/qMvaxZ1Xib | 0.9919697 |
| areyesq | Charlotte @CSoneson is telling us about TreeSummarizedExperiment. That extends SE by adding tree structures to rows and columns of the object. Looks useful for evolution, single cell analyses, etc. #BioC2019 https://t.co/NOSCnPLs61 | 0.9919697 |
| siminaboca | Aedin Culhane @AedinCulhane now presenting on discovery of cancer immune molecular subtypes. She describes the moGSA package for multi-omics gene-set analysis https://t.co/FdKPSImLOX #BioC2019 | 0.9919697 |
| fellgernon |
Denise Scholtens at the Robert Gentleman: “Have you heard about reproducible research? It takes more time, but in the long run, it’s a lot easier for everyone” #BioC2019 #rstats #reproducibility https://t.co/tonEZHEKAL |
0.9919697 |
| ivivek87 | Adding up the framework model i.e CNN based to give an understanding of underlying NN layers of input, hidden and output layers. #bioc2019 https://t.co/Acf6Waabto | 0.9914399 |
Topic 3
| screen_name | text | gamma |
|---|---|---|
| stephaniehicks | @cmirzayi I’m so sorry to hear that Chloe. I do hope you report it to #bioc2019 committee. I know this doesn’t nes help but I can def relate. I’ve faced sim lack/conf comments my entire life & still do today. I’m happy to talk anytime about my exps & how to move past in acad. Just DM me! | 0.9952226 |
| fellgernon |
Wow! I didn’t know this! It’s mind blowing 🤯 Thanks for sharing this @AedinCulhane! Men take 1 yr paternity leave/extension, get more publications. Women take 1 yr, don’t get as many pubs. Hence this mat/pat extension policy hurts on avg more women! #diversity #BioC2019 https://t.co/Svz5UCayj5 |
0.9946292 |
| siminaboca | Robert Castelo @robertclab is looking at RNAseq and proteomics from neonatal dried blood spots (DBS). He’s focusing on preterm infants, especially extremely low gestational age newborns (ELGANs) from whom it would be very challenging to draw peripheral blood. #BioC2019 | 0.9941444 |
| k3yavi | Just arrived at NYC for #BioC2019 . Psyched for 3 days of awesome learning, cheers to @Bioconductor travel fellowship for giving the opportunity. For single cell quantification enthusiasts, come checkout alevin poster in Weiss lobby from 5:30 to 7:30pm on Tuesday. | 0.9941444 |
| fellgernon |
Aedin @AedinCulhane showed the history of women & #diversity in biostatistics & in the @Bioconductor project Wording, quizzes (so women realize they are indeed qualified to attend) & most significantly direct outreach ✅ helped drive up women apps to @ncbi’s hackathon #BioC2019 |
0.9938676 |
| siminaboca | @lievenClement LC said that, despite having really good properties, MsqRob has been difficult to disseminate given the lack of protein summaries and the fact that a mixed model approach only allows for asymptotic inference, without being to calculate degrees of freedom for t test. #BioC2019 | 0.9928543 |
| LeviWaldron1 | I want to state that the behavior described by @cmirzayi after her workshop likely violated the #BioC2019 CoC rules against intellectual bullying. Such behavior reported anonymously or non-anonymously during the conference will be addressed promptly https://t.co/XyD4SgbxJf (1/2) https://t.co/VLgwEzebOi | 0.9919697 |
| KevinRUE67 | Fellow #bioc2019 attendees: if you’re staying a few extra days, an awesome way of moving through the city is the 7-day metro card ($33). One trip is $2.75, in comparison. https://t.co/g78Km9g0z7 | 0.9919697 |
| PayalYokota | Great day at the #bioc2019 @RockefellerUniv today. Saw some #scRNAseq and bumped into some conf posse. Pumped about the 4pm workshops. But, first.. taking a breather to enjoy a delicious mocha @lepainquotidien. https://t.co/w8kBqmTUHi | 0.9919697 |
| mattbrauer | In NYC for #BioC2019 and went to see #Stonewall yesterday–almost 50 years to the day after the resistance was launched. I was surprised at how moving it was, but after all it represents a major turning point in human history. People’s minds can change, though it may take 50 yrs. | 0.9919697 |
Topic 4
| screen_name | text | gamma |
|---|---|---|
| CDSBMexico |
If you are interested in joining our Slack space please do so at https://t.co/lOCIVqWzse ¿Te interesa ser parte de nuestra comunidad? Únete vía https://t.co/lOCIVqWzse FYI most channels are currently in Spanish, but we welcome English-speaking users #BioC2019 #rstats #rstatsES |
0.9941444 |
| cmirzayi | #Bioc2019 Hype post for my workshop this afternoon: Epidemiology for Bioinformaticians. I promise you dogs, DAGs, cats, confounders, an ambulance, toys, GWAS, MR, and a small amount of sleep disruption! Today at 4:10 in Weiss 305. Here’s the cat! https://t.co/KdG7PTg9vK | 0.9935634 |
| areyesq | Ana’s great talk about the integration of COPD and IPF molecular profiling data for easy access for clinicians and wet lab researchers (as a web resource) and bioinformaticians as an R package. #BioC2019 https://t.co/KW0CPVjEPm | 0.9932273 |
| robertclab | JL: used known sex phenotype in GTEx & TCGA to predict that missing phenotype in recount2 https://t.co/Ex0eVN2sVz #BioC2019 now describing the broader problem of predicting genotypes from gene expression data | 0.9924378 |
| fellgernon |
It’s great to see others re-using our work =) See TCGAbiolinks::TCGAquery_recount2() Congrats 🎉 #TCGAbiolinks team on https://t.co/DJyn6wNJHf ^^ ✅ Cites #recount2, #RailRNA and the recount workflow (a version of which I’ll teach at #BioC2019 next week) #rstats @Bioconductor https://t.co/Fy5MaXGhWy |
0.9919697 |
| siminaboca | Really nice setup for the workshops at #bioc2019! Can use Amazon Machine Instances (AMIs) for @rstudio - seems to have minimal lag! Also have the tutorials that folks can follow with/copy paste from at https://t.co/LAwz8IdvnL | 0.9919697 |
| siminaboca | @acolaprico AC also presented DeepBlueR https://t.co/3L0uUR1Ix1, for accessing epigenetics data, and MoonlightR https://t.co/nralZMAMqd for identifying cancer driver genes (oncogenes and tumor suppressors) #BioC2019 | 0.9919697 |
| ivivek87 |
Day 2: @jtleek using recount2 for prediction of missing phenotype using known sex phenotype in GTEx & TCGA App link: https://t.co/BGzPiMeBCR #BioC2019 Slide link: https://t.co/A3T9aKfIU1 |
0.9914399 |
| PayalYokota | Great session about QC’ing those #ATACseq libraries by Haibo and chime ins from Julie and Jianhong. Unfortunately, the R didn’t seem to load libraries for hands-on but I think we all have enough legos to build our own, at home. #BioC2019 | 0.9914399 |
| fellgernon |
Wolfgang Huber @wolfgangkhuber at the Robert Gentleman symposium highlighted how the @Bioconductor project is centered around turning users into developers, biologists into statisticians, statisticians into biologists and much more!! #BioC2019 https://t.co/8hZEtJK2Nb |
0.9914399 |
Topic 5
| screen_name | text | gamma |
|---|---|---|
| dommajeur | #Canicule #chaleur #abeilles #biodiversite #biotech #BioC2019 #bio #biodiversity #ESPUSA #agri #agriculture #charliehebdo #mediapartlive #pollution #pollen #ClimateChangeIsReal #ClimateChange #franceinfo #franceinter #France5 #France #TF1 #Le20hLeMag https://t.co/pqNrlJuLyw | 0.9943973 |
| seandavis12 | Just completed (hopefully) final version of the #bioc2019 workshop materials; 26 workshop packages with 607 dependent packages. All available via #docker image (and volume) and Amazon Machine Image. https://t.co/59hjl9F36i Team of folks involved, not to mention workshop authors. | 0.9941444 |
| LeviWaldron1 | Copy number variation analysis with Bioconductor: Data representation and manipulation, Integrative downstream analysis (eQTL, GWAS, …), Allele-specific CN analysis in cancer. @M2RUseR, @drsehyun, Ludwig Geistlinger. #bioc2019 https://t.co/hVuk5ftFZH | 0.9935634 |
| fellgernon |
Matt @matthewnmccall, @jhubiostat alumni, introducing current @jhubiostat faculty @stephaniehicks at #BioC2019 🙌🏽 Co-founder of the @CorrespondAuth @RLadiesBmore and a force behind #scRNAseq methods & analysis development using @Bioconductor + documention writing w @drisso1893 https://t.co/B0Jzen2ldJ |
0.9928543 |
| mikelove |
My #BioC2019 workshop is 4:10-5pm in Weiss 305. It’s right after @_StuartLee’s workshop at 3:10. plyranges is a great Bioc pkg for range operations w/ dplyr-like syntax, developed by @_StuartLee, @lawremi, @visnut I’ll show using plyranges with tximeta https://t.co/op2AZJdaF8 |
0.9928543 |
| robamezquita | Happy to see that @Bioconductor recognizes that it needs to diversify it’s leadership/advisory boards. It’s tough to admit that out loud, but at #bioc2019 they took a huge positive step forward into the future. Can’t wait to see the new initiatives that will grow from this moment | 0.9928543 |
| siminaboca | @jtleek JL asking what makes primary cancer different from metastatic cancer. Want to make this easy for many people to explore, eg by using recount2 but this uses SRA data, which has mixed or poor quality meta-data. #bioc2019 | 0.9928543 |
| ivivek87 | This is one conference I wish to attend in future. #BioC2019 seem 🔥. Twitter is all I have to learn about it this year. Bring it on. Something I learnt early on. Don’t reinvent wheel if you Jane options, one can work their way through if one digs deep. # @Bioconductor speaks! https://t.co/sabVIm0cyC | 0.9919697 |
| robamezquita | @RevDocGabriel Big fan of yours thanks to your question - I see no reason why we should be exclusivist regarding creating artificial barriers to entry for new developers. Lots of great statistical talent that is untapped that could make great genomic analysis software #bioc2019 | 0.9919697 |
| annaquagli | That 😊feeling when you are leaving the longest and coldest ❄ nights in Melbourne to go to summer☀in NY! Exciting week ahead attending #bioc2019 and visiting @jhubiostat for the first time! #rstatstour | 0.9914399 |
Topic 6
| screen_name | text | gamma |
|---|---|---|
| fellgernon |
Took me a while, but I can now run Install https://t.co/5s6anvxrFT docker login docker run hello-world Next docker pull bioconductor/release_base2 docker run -p 8787:8787 -e PASSWORD=bioc |
0.9938676 |
| AedinCulhane | Truly excellent #bioc2019. I was so impressed with the high quality badges designed by @jotsetung and @_schifferl. The local organizers Tomas Carroll @RockefellerUniv, Kelly Ruggles @nyuniversity and @LeviWaldron1 went the extra mile P<0.05 https://t.co/JIegSGHZsr | 0.9938676 |
| siminaboca | Benjamin Haibe-Kains @bhaibeka now presenting on robust biomarker discovery from cancer pharmacogenomic data. Can use his group’s packages https://t.co/o5dqAR8nbZ to get datasets and analyze the data. He’s focused on robust biomarkers that are not platform-dependent #BioC2019 | 0.9935634 |
| CDSBMexico |
Congrats @AnaBetty2304 @josschavezf1 @cmvaldezcordova for your presentations at #BioC2019 today! 🇲🇽 💪🏽 Soon near you BiocManager::install(#PulmonDB #regutools #histoneSig) 😁 ¡Felicidades Ana, Joselyn y César por sus presentaciones hoy en el congreso de @Bioconductor! 👏🏽 🙌🏽 https://t.co/sbv9u823sL |
0.9935634 |
| PeteHaitch | Love seeing in the talks already the strong sense of community in @Bioconductor software development. Lots of people credited and thanked, people working together to create great tools, and building upon the foundational work of the core team #BioC2019 #rstats | 0.9932273 |
| Bioconductor | @Bioconductor #bioc2019 in my (M. Morgan) thanks this AM I meant to include a special shout-out to Tom Carroll @Rockefeller_BRC and Kelly Ruggles @kruggle for amazing and heroic work as local organizers; the event has been exceptionally popular and successful due to their effort | 0.9932273 |
| AedinCulhane | Thank you, organizers, for an excellent #bioc2019. Beautiful location, great facilities, stimulating science, great talks, workshops and people. Especially, thanks to local hosts Thomas Carroll, Kelly Ruggles and @LeviWaldron1, Matthew McCall https://t.co/6gnh6oqOhX | 0.9932273 |
| siminaboca | @lievenClement LC very clearly explaining the mass spec workflow and the challenges with proteomics quantitation and identification. Need to look up MS2 peaks in databases. “Proteomics people are addicted to big databases.” #BioC2019 https://t.co/rAXds6QWVD | 0.9928543 |
| robertclab | Today I’ll be giving a talk on technical aspects of this work at the @Bioconductor meeting #bioc2019 and on Sept 20th @CostaCoto will be giving a talk at the @JENS_Congress on its contribution to our understanding of the postnatal effects of the fetal inflammatory response 7/7 | 0.9928543 |
| k3yavi | #BioC2019 My first bioconductor conference and it couldn’t have been better. I met with so many brilliant researchers and exchanged ideas. I never realized how the idea of forming a community of similar mind researchers can be so powerful to move science forward, it’s incredible! | 0.9928543 |
9 Software
Software mentioned in Tweets with links to GitHub, BitBucket, Bioconductor or CRAN.
Session info
## R Under development (unstable) (2021-02-18 r80027)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.1 LTS
##
## Matrix products: default
## BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices datasets utils methods base
##
## other attached packages:
## [1] fs_1.5.0 here_1.0.1 kableExtra_1.3.1
## [4] knitr_1.30 magick_2.5.2 webshot_0.5.2
## [7] viridis_0.5.1 viridisLite_0.3.0 wordcloud_2.6
## [10] RColorBrewer_1.1-2 ggraph_2.0.4 ggrepel_0.8.2
## [13] ggplot2_3.3.2 topicmodels_0.2-11 tidytext_0.2.6
## [16] igraph_1.2.6 stringr_1.4.0 purrr_0.3.4
## [19] forcats_0.5.0 lubridate_1.7.9.2 tidyr_1.1.2
## [22] dplyr_1.0.2 rtweet_0.7.0 BiocManager_1.30.10
##
## loaded via a namespace (and not attached):
## [1] httr_1.4.2 tidygraph_1.2.0 jsonlite_1.7.2 assertthat_0.2.1
## [5] askpass_1.1 highr_0.8 stats4_4.1.0 renv_0.12.3
## [9] yaml_2.2.1 slam_0.1-48 pillar_1.4.7 lattice_0.20-41
## [13] glue_1.4.2 digest_0.6.27 polyclip_1.10-0 rvest_0.3.6
## [17] colorspace_2.0-0 plyr_1.8.6 htmltools_0.5.0 Matrix_1.2-18
## [21] tm_0.7-8 pkgconfig_2.0.3 scales_1.1.1 processx_3.4.5
## [25] tweenr_1.0.1 ggforce_0.3.2 tibble_3.0.4 openssl_1.4.3
## [29] generics_0.1.0 farver_2.0.3 ellipsis_0.3.1 withr_2.3.0
## [33] cli_2.2.0 NLP_0.2-1 magrittr_2.0.1 crayon_1.4.1
## [37] ps_1.5.0 evaluate_0.14 tokenizers_0.2.1 janeaustenr_0.1.5
## [41] fansi_0.4.1 SnowballC_0.7.0 MASS_7.3-53 xml2_1.3.2
## [45] tools_4.1.0 lifecycle_0.2.0 munsell_0.5.0 callr_3.5.1
## [49] compiler_4.1.0 rlang_0.4.9 grid_4.1.0 rstudioapi_0.13
## [53] labeling_0.4.2 rmarkdown_2.6 gtable_0.3.0 curl_4.3
## [57] reshape2_1.4.4 graphlayouts_0.7.1 R6_2.5.0 gridExtra_2.3
## [61] utf8_1.1.4 rprojroot_2.0.2 modeltools_0.2-23 stringi_1.5.3
## [65] parallel_4.1.0 Rcpp_1.0.5 vctrs_0.3.5 tidyselect_1.1.0
## [69] xfun_0.19