iSEE workshop at Bioc2019!
The iSEE package will be presented as a workshop at the Bioconductor 2019 conference in New York City!
What to expect
Charlotte Soneson and I will be conducting a workshop that demonstrates the many features and usages of the iSEE package.
With the help of the iSEE team, we have prepared a vignette demonstrating the integration of the package functionality into an example single-cell RNA-sequencing workflow.
The workshop was designed to cater for the needs of users at all levels of familiarity with R and the Bioconductor project.
Workshop content
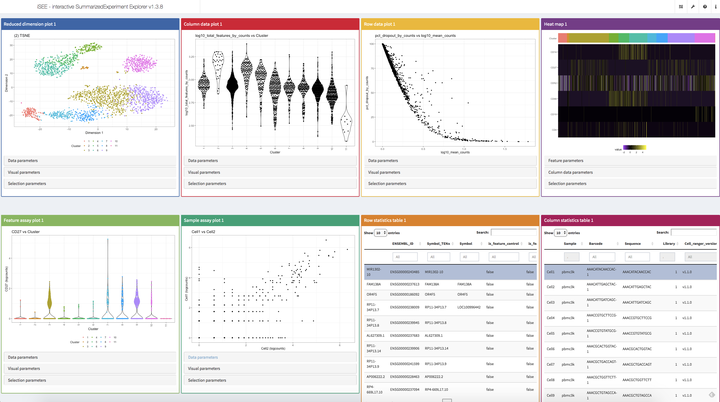
We will first take time to introduce users to the SummarizedExperiment and SingleCellExperiment classes in the context of a single-cell RNA-sequencing workflow. Next, we will walk participants through the various components of the graphical user interface (GUI). Finally, we will demonstrate how individual instances of iSEE applications can be preconfigured to save time and share insights with others.
Throughout the workshop, we will showcase key features of the package that make it a great companion for interactive and reproducible analysis for a variety of genomics data modalities:
- A multi-panel layout to simultaneously visualise multiple aspects of the dataset.
- Point selections using shiny brushes or lasso polygons drawn by consecutive clicks.
- Multiple concurrent selections supported in versions ≥ 1.3.8.
- Dynamic linking between plot and table panels, allowing users to transmit information across multiple panels via point selection.
- Dynamic addition, removal, reordering, and resizing of panels.
- Automatic tracking, storage, and rendering of the exact R code in a shinyAce editor to allow regeneration of all visible plots in a given application instance.
- Intelligent density-dependent downsampling of data points shown for large data sets.
- Custom colour scale specification for individual and groups of discrete and continuous variables.
- Construction of bespoke interactive guided tours using rintrojs.
- Dynamic detection of discrete and continuous covariates, switching individual plot panels between scatter plots, violin plots, and “square” plots.
- Ability for users to programmatically define additional custom panels based on any number of user-defined functions performing on-the-fly calculations.
- Ability to preconfigure app instances, including point selections and state of collapsible panels, to launch individual app instances in any predefined state.
- Retrieval of gene annotations on the fly by using either user-defined functions or the built-in annotateEntrez or annotateEnsembl functions.
- Hands-free control of the app using voice recognition by annyang in versions ≥ 1.3.2.
Where and when to find us
This workshop will take place during the workshop sessions 6a and 6b on Wednesday June 26 between between 15:10 and 17:00.
You can also come and find us between sessions if you have more questions!
Access to the workshop materials
The workshop materials are being prepared as the vignette of a freely available R package hosted on a GitHub repository.
In addition, workshops will become part of a book similar to the Bioconductor 2018 Workshop Compilation.
We look forward to seeing participants at the workshop!